Abstract
Restriction of HIV-1 in myeloid-lineage cells is attributed in part to the nucleotidase activity of the SAM-domain and HD-domain containing protein (SAMHD1), which depletes free nucleotides, blocking reverse transcription. In the same cells, the Vpx protein of HIV-2 and most SIVs counteracts SAMHD1. Both Type I and II interferons may stimulate SAMHD1 transcription. The contributions of SAMHD1 to retroviral restriction in the central nervous system (CNS) have been the subject of limited study. We hypothesized that SAMHD1 would respond to interferon in the SIV-infected CNS but would not control virus due to SIV Vpx. Accordingly, we investigated SAMHD1 transcript abundance and association with the Type I interferon response in an SIV model. SAMHD1 transcript levels were IFN responsive, increasing during acute phase infection and decreasing during a more quiescent phase, but generally remaining elevated at all post-infection time points. In vitro, SAMHD1 transcript was abundant in macaque astrocytes and further induced by Type I interferon, while IFN produced a weaker response in the more permissive environment of the macrophage. We cannot rule out a contribution of SAMHD1 to retroviral restriction in relatively non-permissive CNS cell types. We encourage additional research in this area, particularly in the context of HIV-1 infection.
Similar content being viewed by others
Introduction
The sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1) is a 72-kD protein originally characterized as the human ortholog1 of a mouse gamma interferon-induced gene2. SAMHD1 was later found to have mutations associated with Aicardi-Goutières syndrome3. More recently, SAMHD1 was identified as a host restriction factor for HIV-14,5. Restriction factors block or interfere with one or more stages of the viral life cycles and include but are not limited to members of the APOBEC cytidine deaminase family, tetherin, TRIM5alpha and cellular microRNAs6,7,8.
SAMHD1 in its activated state depletes the cellular pool of deoxynucleotides through deoxynucleoside triphosphohydrolase activity9,10,11. Depleting the dNTPs needed to build genomic DNA, SAMHD1 thus exerts antiviral restriction by slowing or stopping the reverse transcription step of HIV-1 replication. Consistent with the antiviral role of SAMHD1, cells from individuals with Aicardi-Goutières syndrome have heightened susceptibility to HIV-1 infection12. Exonuclease activity of SAMHD1 has also been described13. The Vpx accessory proteins of HIV-2 and SIV target SAMHD1 for interaction with the E3 ubiquitin ligase complex14, resulting in proteasome-mediated degradation and counteraction of the restriction activity4,5,15. The Vpr protein of some primate lentiviruses can also orchestrate SAMHD1 degradation16.
SAMHD1 expression and its control have been examined extensively in certain cell types. SAMHD1 has been found to be relatively highly expressed in cells that are restrictively permissive to HIV infection, such as macrophages and dendritic cells of myeloid cell origin17 or resting CD4+ T-cells18. In contrast, some permissive cells may display lower levels of SAMHD1. Transcription of SAMHD1 can be stimulated by interferon gamma2,19. There is also evidence for stimulation by Type I interferons12,20,21,22,23, but evidence against interferon stimulation has also been presented20,24,25. At the same time, in some cells, SAMHD1 may be part of a feedback system, as it has been reported to antagonize the expression of certain Type I IFN-stimulated genes26,27, strikingly in monocyte-derived dendritic cells28. However, some myeloid-lineage cells and resting CD4+ T-cells from individuals with SAMHD1-inactivating mutations supported robust infection in vitro12,29, suggesting that the observed anti-IFN activity of SAMHD1 is cell type specific or that its ablation is insufficient to allow the interferon response to protect against HIV-1 infection. Differential transcript-level expression of SAMHD1 has been reported in cells of one cohort of HIV-1 elite suppressors30, but not in two others31,32.
In contrast with the periphery, relatively little is known about SAMHD1 levels during retroviral infection of the brain and progression to the HIV-associated neurocognitive disorders (HAND). Neurologic correlates of HIV infection have been observed since the earliest days of the pandemic33,34,35 and may even occur despite successful virologic control36,37,38. Some pharmacologic agents may contribute to HAND39 by inducing oxidative stress. Infiltrating macrophages and resident microglia are productively infected40. Astrocytes, the most abundant cells in the brain, are also infected41, albeit rarely productively in vivo. In one study of SAMHD1 in CNS cells, interferon alpha transiently increased SAMHD1 production in immortalized human stem cell-derived astrocytes23. In another, astrocytes were found to have higher levels of SAMHD1 than microglia, which are relatively permissive to infection42. Host miR-181a was found to regulate SAMHD1 in vitro42, although miRNA association with SAMHD1 levels was not apparent in vivo43.
Since SAMHD1 transcription and Type I interferon responsiveness in the retrovirus-infected CNS are unknown, we hypothesized that SAMHD1 transcript levels might respond to SIV infection, but without affecting viral replication due to the action of SIV Vpx. We further hypothesized that macaque astrocytes, which are relatively refractory to infection and host only low levels of viral replication, would display high levels of SAMHD1 transcription (as now also reported for human astrocytes42). We therefore examined transcription of SAMHD1 in thalamus in an SIV model44 in which an acute infection phase of ten to 14 days includes peak cytokine responses45 and is followed by a latent or asymptomatic phase. Recapitulation of markers of human disease progression is accompanied by AIDS and CNS disease by three months post-infection in >90% of cases, with sustained inflammation and other factors contributing to neurodegenerative pathologies46. SAMHD1 transcript was also measured in primary macaque astrocytes infected with SIV and/or treated with interferon beta, the Type I interferon most important in the CNS response to viral replication.
Our results indicate that SAMHD1 mRNA expression in thalamus mirrors the rise of proinflammatory cytokine expression during acute infection and appears to remain elevated during asymptomatic infection and into disease progression. In vitro, primary macrophages and astrocytes did not undergo significant changes in SAMHD1 expression during the course of long-term infection with HIV or SIV, although early changes were observed in SIV-infected astrocytes. However, treatment of macrophages and especially astrocytes with exogenous interferon-beta resulted in significant fold changes in SAMHD1.
Results
Regulation of SAMHD1 transcript in CNS during retroviral infection
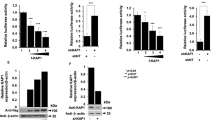
Thalamus RNA samples from 58 subjects (Table 1) were obtained and subjected to quantitative PCR. With reference to the average of two housekeeping genes, GAPDH and beta actin and compared with levels in six uninfected control animals, SAMHD1 was significantly increased during acute infection (4, 7 and 10 dpi) (Fig. 1). Indeed, an increase over baseline was maintained at later time points in all but a small number of subjects, even during the “latent” or “asymptomatic” phase that includes 14 and 21 dpi. This increase was nominally significant at 56 dpi, but this should be viewed in the context of the broader trend towards increased expression.
Brain SAMHD1 transcript load during SIV infection.
SAMHD1 transcript increased in thalamus during SIV infection, with peak expression corresponding with acute and late-stage infection. SAMHD1 mRNA was quantitated by qPCR, with normalization to the average of beta actin and GAPDH. **p < 0.01, *p < 0.05.
SAMHD1 and the Type I interferon response
Based upon reports in the literature and our experience with measuring host antiviral responses45, we postulated that the observed increases in SAMHD1 are part of the Type 1 interferon antiviral program. To investigate this possibility, we first used a bioinformatics approach to confirm that the macaque SAMHD1 promoter sequence, like that of human, contains interferon stimulated response elements (ISREs). MatInspector predicted ISREs in both human and animal sequences (Table 2). Next, we compared expression of SAMHD1 and the interferon stimulated gene Mx1 (Fig. 2A). A strong correlation between the two was observed. This finding is consistent with Type I interferon responsiveness of both genes.
Relationship of SAMHD1 transcription, Type I interferon response and disease severity.
There was a significant positive correlation of SAMHD1 transcript and level of MX1, an indicator of the Type I interferon response (A). Comparing Mx1 (B) and SAMHD1 (C) transcript with disease severity as assigned by pathology (none, mild, moderate, or severe), the association of Mx1 with disease appeared stronger than the association of SAMHD1 and disease. Transcript abundance was normalized to the average of beta actin and GAPDH; hence, the smaller the “dCq” value, the greater the abundance.
SAMHD1 transcript during lentiviral brain disease development
In the rapid model, disease develops by the 42 and 56 day post-inoculation time points. While Mx1 was correlated with CNS disease severity (Fig. 2B), SAMHD1 displayed a weaker correlation with disease (Fig. 2C) at these time points. A trend towards greater expression of SAMHD1 in severe disease was partially offset by several “non-responsive” animals with lower SAMHD1 expression despite high MX1 response (Fig. 2C). As we hypothesized, however, higher levels of SAMHD1 expression did not associate overall with lower viral loads in brain tissue (Fig. 3). Both MX1 and SAMHD1 expression were positively correlated with severity of CNS disease, consistent with a runaway immune response that, in acute phase infection, might be protective, but at later stages becomes damaging.
SAMHD1 regulation in monocyte-derived macrophages
To follow up on the in vivo findings consistent with Type I interferon responsiveness of SAMHD1 in brain, we next sought to determine which cell types might be responsible for SAMHD1 regulation in the CNS. We assayed SAMHD1 expression in several relevant cell types in vitro, in response to interferon beta (IFNB1) treatment or infection. The first cell type assayed was the human monocyte-derived macrophage. We studied human macrophages because of their relative availability at the time of these experiments. Following infection, monocyte-lineage cells enter the brain within days, seeding CNS viral infection. During progression to disease, perivascular and parenchymal macrophages support productive infection and release damaging proinflammatory cytokines.
In human monocyte-derived macrophages, differentiated by response to macrophage colony stimulating factor (M-CSF), Mx1 was upregulated rapidly and significantly as early as 2 hours post-treatment with recombinant IFNB1 (Fig. 4A). In contrast, TNFalpha did not trigger significant changes in Mx1. Interferon beta resulted in later upregulation of SAMHD1, beginning between 8 and 12 hours post-treatment (Fig. 4B). This increase was significant at 12 and 24 hours. It is unclear why the effect was delayed for SAMHD1 compared with Mx1. Possibly, the interferon effect is indirect, requiring initiation of a secondary signaling cascade. TNFalpha treatment resulted in apparently lower SAMHD1 expression. However, this trend did not reach significance.
SAMHD1 and IFNB1 in primary macrophages.
M×1 of primary human monocyte-derived macrophages responded rapidly and strongly to 100 U/mL treatment with recombinant IFNB1, but not to TNFalpha (20 ng/mL, (A)). SAMHD1 also responded to IFNB1, but the increase was detectable and significant only by 12 hours post-treatment (B). HIV-1 (BaL) infection of primary monocyte-derived macrophages yielded an apparent increase of SAMHD1 levels by 7 and 10 days post-infection, but this increase was not statistically significant (NS) (C). ***p < 0.001, *p < 0.05.
In parallel, we assessed the effect of HIV infection on macrophages. Cells were infected at a multiplicity of infection of 0.05 and samples were harvested at time points including two, seven, nine, 14 and 21 days post-infection. Productive infection was confirmed by p24 ELISA (not shown). Despite an apparent trend towards increased SAMHD1 abundance at seven and nine dpi, none of the changes was significant (Fig. 4C). Given the significant increase in SAMHD1 in interferon beta-treated cells, this result might seem counterintuitive. Would infected or exposed cells not generate interferon and thus render themselves and surrounding cells less susceptible to infection or replication? There are several explanations we can offer. First, macrophage innate responses to HIV infection may be turned off or subverted by viral products or specific cellular responses to viral products47. As cited above, according to several reports, SAMHD1 itself can dampen IFN responses under certain circumstances. Second, the timing of the experimental exposure to recombinant interferon may have led to negative feedback by cellular factors such as miRNAs48, influencing results. Finally, replicating in vitro the microenvironment of the brain during SIV infection may be a considerable challenge.
SAMHD1 regulation in primary astrocytes
The second cell type we tested was the primary astrocyte. To take advantage of a unique resource—rhesus adult primary astrocytes—this experiment was performed with SIV/17E-Fr infection, not HIV. The most abundant cell type in the brain, astrocytes, are readily infected in vitro49. They are also infected in vivo, but incompletely characterized blocks to replication usually prevent productive infection. By sheer numbers, however, astrocytes comprise the most abundant, if largely inactive, lentiviral reservoir in the CNS.
Astrocytes infected or not with SIV/17E-Fr at a multiplicity of infection of 0.1 were then exposed or not to 100 units of recombinant macaque IFNB1 per milliliter of culture medium, creating four experimental conditions. We compared the change from pre-infection at post-treatment time points of two, seven, 14 and 21 days, with the untreated, uninfected condition as the reference at each time point. At two days post-treatment, SAMHD1 levels were significantly upregulated in all interferon and infection conditions, including an approximately eight-fold upregulation in response to interferon (Fig. 5). Thereafter, SAMHD1 remained upregulated in the presence of exogenous interferon but not in the SIV alone condition. The large fold change of SAMHD1 in astrocytes compared with macrophages suggests, with the caveats noted above, that astrocytes may contribute a larger portion of the observed upregulation of SAMHD1 transcript in brain. Furthermore, it is interesting to note the comparatively greater response of SAMHD1 in the astrocyte, generally considered a non-permissive cell type, versus the permissive macrophage. However, since the astrocytes used in these in vitro experiments do support viral replication, additional work will be needed to elucidate the details of the role of SAMHD1 in enforcing viral latency in astrocytes in vivo.
Primary astrocyte SAMHD1 responds to SIV infection and IFNB1.
Quantitative PCR results, normalized to the average of beta actin and GAPDH, indicated that SIV infection (SIV 17E-Fr) and IFNB1 treatment of primary rhesus astrocytes resulted in significant upregulation of SAMHD1 by 48 hours post infection and at all subsequent time points examined. SIV infection alone stimulated apparently significant upregulation at two and 14 days post infection. ***p < 0.001, **p < 0.01, *p < 0.05.
Discussion
We report that SAMHD1 expression is significantly upregulated in thalamus during retroviral infection and tracks with the Type I interferon response as represented by MX1. Our in vitro results suggest that, in part, this upregulation of SAMHD1 may be driven by the strong response of astrocyte SAMHD1 to Type I interferon—specifically IFNB1—as well as a comparatively minor and short-lived response to retrovirus exposure. We cannot rule out that this high level of SAMHD1 expression may help to protect astrocytes from retrovirus infection, even in the case of Vpx-expressing SIV. Although the contribution of macrophages is substantially less clear due to contradictory results in the literature, our findings suggest that macrophages may also contribute an augmented complement of SAMHD1 following IFNB1 exposure.
Despite the upregulation of SAMHD1 in thalamus during SIV infection, SAMHD1 transcript levels do not correlate negatively with viral load. Thus, the arms race against the retrovirus is lost overall, likely in part due to SIV counteraction of SAMHD1 protein in infected cells. This contrasts with our results on HIV-1 in the periphery, where, in a study of PBMC of HIV-1 elite suppressors and chronic progressors, a negative correlation of viral load with MX1 and SAMHD1 was observed in viremic individuals31. It will be interesting to follow up on these findings in HIV-infected brain.
Strategies to upregulate SAMHD1, for example during “shock and kill” reactivation therapy50, could supplement the actions of traditional antiviral drugs and further prevent the spread of any newly produced virus. Transcriptional regulation is of course not the only determinant of protein activity. One form of post-transcriptional regulation is microRNA-mediated repression: regulation of the SAMHD1 transcript may include post-transcriptional blocks such as miRNA-mediated degradation or translational attenuation. As we reported recently43 and at the 2013 Conference on Retroviruses and Opportunistic Infections51, miRNAs with seed sequence matches to SAMHD1 that appear to bind to the transcript include miRs-34a, -125b, -150, -155 and the -181 family. This effect has recently been confirmed for miR-181a and miR-15542 and may occur for other miRNAs as well7. The finding that host miRNAs contribute to SAMHD1 regulation could form the basis of a SAMHD1-enhancing strategy as mentioned above. Protein-level activity is also governed by post-transcriptional factors including phosphorylation state52,53 and multimerization54. Blocking Type I interferon signaling in myeloid and plasmacytoid dendritic cells allowed the cells to become permissive to HIV-1 infection55 and the antiviral form of SAMHD1, without phosphorylation on residue Threonine 592, is promoted by interferon53. Accordingly, the phosphorylation and multimerization states of SAMHD1 should also be investigated further in the CNS.
Methods
Animal studies and ethics statement
Studies of animal materials were conducted using archived samples from previous, concluded pigtailed macaque studies involving dual-inoculation with an immunosuppressive SIV swarm and a neurotropic clone56; no new animal subjects were involved in this project. Characteristics of the subjects are outlined in Table 1. In addition to uninfected controls, post-infection time points examined were 0, 4, 7, 10, 14, 21, 42, 56 and 84 dpi. All animal studies were approved by the Johns Hopkins University Institutional Animal Care and Use Committee and conducted in accordance with the Weatherall Report, the Guide for the Care and Use of Laboratory Animals and the USDA Animal Welfare Act.
Macrophage differentiation
Peripheral blood mononuclear cells (PBMCs) were isolated from de-identified blood products by standard Percoll protocol as described previously57,58. PBMCs were cultured at a density of 4 × 106 cells per well in 12-well plates or 2 × 106 cells per well in 24-well plates in macrophage differentiation medium (MDM) with 20% serum [MDM20: Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen), 20% human serum (GemCell), 2 mM L-Glutamine (Sigma), 10 mM HEPES (Gibco), 10 mM sodium pyruvate (Sigma), 50 ng/ml M-CSF (R&D) and 2 mg/ml Gentamicin (Gibco)] for one week, with half-volume re-feeding at three days post-plating. Note that human serum lots were selected for minimal pyrogen activity and tested in cell culture before use. After seven days, supernatants were removed and adherent macrophages were washed with PBS. Medium was replaced with MDM with 10% human serum.
Macrophage infection, harvesting and RNA isolation
Differentiated macrophages in 12-well plates were infected with HIV-1-BaL (50 ng p24) for 4–6 hours and virus was removed with extensive washes with PBS. Infected and uninfected cells were harvested in mirVana RNA lysis buffer (Ambion/Life Technologies) day 0 and at 2, 7, 9, 14 and 21 dpi and stored at −80 °C. Supernatants were also collected and stored at −80 °C.
Short-term macrophage cytokine treatment
Macrophages were sham treated or treated with 100 units/mL of recombinant human IFN-beta or 20 ng/mL TNF-alpha (PBL Interferon Source, now PBL Assay Science, Piscataway, NJ). Lysates were collected at 2, 4, 8, 12 and 24 hours post-treatment as described above.
Astrocyte infection with SIV
Primary macaque astrocytes were purchased from Cambrex and cultured in astrocyte growth media with supplements (Lonza) as described previously41. Twenty-four hours prior to infection, astrocyte medium was changed to DMEM with 10% fetal bovine serum. Cells were infected at moi = 0.05 with SIV/17E-Fr59. At six hours post-infection, cells were washed five times with PBS and re-fed with fresh medium. The next day, cells were treated or not with 100 units/mL of recombinant macaque IFN-beta (PBL Assay Science, Piscataway, NJ). Cells were half re-fed twice weekly with medium containing the cytokine as appropriate. Astrocyte lysates at the specified time points were collected in mirVana lysis buffer and stored at −80 °C until RNA purification.
RNA isolation from tissue and cell culture
Thalamus tissue from the rapid Macaca nemestrina model of HIV CNS disease was obtained after long-term maintenance at −80 C. RNA was extracted with a modified Trizol protocol with column clean-up as previously described60. 30–50 mg frozen tissue was placed into 1 mL Trizol (Invitrogen, Grand Island, NY) in 2 mL screw-cap tubes and one tube of Lysing matrix D (MP Biomedicals, Solon, OH) was added. Tissue was disrupted for 2 × 30 s using a desktop bead beater with five minutes on ice interspersed. Tubes were spun to collect beads and homogenate and 200 μl chloroform was added. Tubes were firmly capped and shaken vigorously for 1 min, then centrifuged at 12,000 × g in an Eppendorf C2415 centrifuge for 15 min at 4 °C. The aqueous phase was transferred to a new tube, with care taken not to disturb the interphase. 1.25 volumes 100% ethanol was added. Sample was applied to mirVana miRNA isolation kit columns for cleanup (Ambion/Life Technologies).
Total RNA was extracted from frozen cell culture samples using the mirVana miRNA isolation kit (Ambion/Life Technologies), following the manufacturer’s protocol. RNA for each experiment was isolated together to avoid batch effects. RNA concentration of all samples was assessed by NanoDrop and integrity of astrocyte RNA and several additional samples was assessed by BioAnalyzer. All RNA was stored at −80 °C.
Quantitative PCR
cDNA was generated using the High-Capacity cDNA Reverse Transcription kit (Ambion/Life Technologies, #4368814) and manufacturer’s protocol. Pre-designed TaqMan Assays (Applied Biosystems/Life Technologies) were used to quantitate SAMHD1, Mx1, GAPDH and ACTB from human (Hs00210019_m1, Hs00895608_m1, Hs02758991_g1 and Hs99999903_m1, respectively) and macaque (Rh02869978_m1, Rh02842279_m1, Rh02621745_g1 and Rh03043379_gH, respectively). Amplification reactions were performed with the CFX96 optical system (BioRad). Delta-delta Cq-based normalization was to the average of housekeeping genes GAPDH and ACTB (Delta Cq) and the average of control, uninfected samples (Delta-delta Cq).
Data analysis, statistical methods and figures
Data processing and analysis were conducted using tools from Microsoft Excel, GraphPad Prism and XLStat. Analysis of Variance (ANOVA) was used to calculate p values. Significance of correlations was calculated as the two-sided probability value of the Pearson correlation coefficient.
Additional Information
How to cite this article: Buchanan, E. L. et al. SAMHD1 transcript upregulation during SIV infection of the central nervous system does not associate with reduced viral load. Sci. Rep. 6, 22629; doi: 10.1038/srep22629 (2016).
References
Li, N., Zhang, W. & Cao, X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol Lett 74, 221–224 (2000).
Lafuse, W. P., Brown, D., Castle, L. & Zwilling, B. S. Cloning and characterization of a novel cDNA that is IFN-gamma-induced in mouse peritoneal macrophages and encodes a putative GTP-binding protein. J Leukoc Biol 57, 477–483 (1995).
Crow, Y. J. et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 38, 917–920 (2006).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011).
Malim, M. H. & Bieniasz, P. D. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med 2, a006940 (2012).
Swaminathan, G., Navas-Martin, S. & Martin-Garcia, J. MicroRNAs and HIV-1 infection: antiviral activities and beyond. J Mol Biol 426, 1178–1197 (2014).
Zheng, Y. H., Jeang, K. T. & Tokunaga, K. Host restriction factors in retroviral infection: promises in virus-host interaction. Retrovirology 9, 112 (2012).
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011).
Powell, R. D., Holland, P. J., Hollis, T. & Perrino, F. W. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem 286, 43596–43600 (2011).
Lahouassa, H. et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13, 223–228 (2012).
Berger, A. et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog 7, e1002425 (2011).
Beloglazova, N. et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem 288, 8101–8110 (2013).
Ahn, J. et al. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem 287, 12550–12558 (2012).
Laguette, N. et al. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217 (2012).
Lim, E. S. et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 (2012).
Kim, B., Nguyen, L. A., Daddacha, W. & Hollenbaugh, J. A. Tight interplay among SAMHD1 protein level, cellular dNTP levels and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem 287, 21570–21574 (2012).
Baldauf, H. M. et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18, 1682–1687 (2012).
Taylor, G. A. et al. Identification of a novel GTPase, the inducibly expressed GTPase, that accumulates in response to interferon gamma. J Biol Chem 271, 20399–20405 (1996).
St Gelais, C. et al. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105 (2012).
Carlow, D. A., Teh, S. J. & Teh, H. S. Specific antiviral activity demonstrated by TGTP, a member of a new family of interferon-induced GTPases. J Immunol 161, 2348–2355 (1998).
Harper, M. S. et al. IFN-alpha treatment inhibits acute Friend retrovirus replication primarily through the antiviral effector molecule Apobec3. J Immunol 190, 1583–1590 (2013).
Cuadrado, E. et al. Chronic exposure of astrocytes to interferon-alpha reveals molecular changes related to Aicardi-Goutieres syndrome. Brain 136, 245–258 (2013).
Goujon, C. et al. Evidence for IFNalpha-induced, SAMHD1-independent inhibitors of early HIV-μμnfection. Retrovirology 10, 23 (2013).
Dragin, L. et al. Interferon block to HIV-1 transduction in macrophages despite SAMHD1 degradation and high deoxynucleoside triphosphates supply. Retrovirology 10, 30 (2013).
Kretschmer, S. et al. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis 74, e17 (2014).
Zhang, R., Bloch, N., Nguyen, L. A., Kim, B. & Landau, N. R. SAMHD1 restricts HIV-1 replication and regulates interferon production in mouse myeloid cells. PLoS One 9, e89558 (2014).
Puigdomenech, I., Casartelli, N., Porrot, F. & Schwartz, O. SAMHD1 restricts HIV-1 cell-to-cell transmission and limits immune detection in monocyte-derived dendritic cells. J Virol 87, 2846–2856 (2013).
Descours, B. et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87 (2012).
Riveira-Munoz, E. et al. Increased expression of SAMHD1 in a subset of HIV-1 elite controllers. J Antimicrob Chemother 69, 3057–3060 (2014).
Buchanan, E. L., McAlexander, M. A. & Witwer, K. W. SAMHD1 expression in blood cells of HIV-1 elite suppressors and viraemic progressors. J Antimicrob Chemother 70, 954–956 (2014).
Abdel-Mohsen, M. et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 10, 106 (2013).
Zhou, L. & Saksena, N. K. HIV Associated Neurocognitive Disorders. Infect Dis Rep 5, e8 (2013).
Spudich, S. HIV and neurocognitive dysfunction. Curr HIV/AIDS Rep 10, 235–243 (2013).
Lindl, K. A., Marks, D. R., Kolson, D. L. & Jordan-Sciutto, K. L. HIV-associated neurocognitive disorder: pathogenesis and therapeutic opportunities. J Neuroimmune Pharmacol 5, 294–309 (2010).
Clifford, D. B. & Ances, B. M. HIV-associated neurocognitive disorder. Lancet Infect Dis 13, 976–986 (2013).
Alfahad, T. B. & Nath, A. Update on HIV-associated neurocognitive disorders. Curr Neurol Neurosci Rep 13, 387 (2013).
McArthur, J. C., Steiner, J., Sacktor, N. & Nath, A. Human immunodeficiency virus-associated neurocognitive disorders: Mind the gap. Ann Neurol 67, 699–714 (2010).
Akay, C. et al. Antiretroviral drugs induce oxidative stress and neuronal damage in the central nervous system. J Neurovirol 20, 39–53 (2014).
Koppensteiner, H., Brack-Werner, R. & Schindler, M. Macrophages and their relevance in Human Immunodeficiency Virus Type I infection. Retrovirology 9, 82 (2012).
Overholser, E. D. et al. Expression of simian immunodeficiency virus (SIV) nef in astrocytes during acute and terminal infection and requirement of nef for optimal replication of neurovirulent SIV in vitro. J Virol 77, 6855–6866 (2003).
Pilakka-Kanthikeel, S. et al. Sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1)-facilitated HIV restriction in astrocytes is regulated by miRNA-181a. J Neuroinflammation 12, 66 (2015).
Witwer, K. W., Buchanan, E. L., Myers, S. L. & McAlexander, M. A. miRNAs and SAMHD1 regulation in vitro and in a model of HIV CNS disease. J Neuroinflammation 12, 159 (2015).
Beck, S. E. et al. Paving the path to HIV neurotherapy: Predicting SIV CNS disease. Eur J Pharmacol 759, 303–312 (2015).
Witwer, K. W. et al. Coordinated regulation of SIV replication and immune responses in the CNS. PLoS One 4, e8129 (2009).
Meulendyke, K. A. et al. Elevated Brain Monoamine Oxidase Activity in SIV- and HIV-associated Neurological Disease. J Infect Dis 210, 904–912 (2014).
Yan, N., Regalado-Magdos, A. D., Stiggelbout, B., Lee-Kirsch, M. A. & Lieberman, J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 11, 1005–1013 (2010).
Witwer, K. W., Sisk, J. M., Gama, L. & Clements, J. E. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol 184, 2369–2376 (2010).
Overholser, E. D., Babas, T., Zink, M. C., Barber, S. A. & Clements, J. E. CD4-independent entry and replication of simian immunodeficiency virus in primary rhesus macaque astrocytes are regulated by the transmembrane protein. J Virol 79, 4944–4951 (2005).
Archin, N. M. & Margolis, D. M. Emerging strategies to deplete the HIV reservoir. Curr Opin Infect Dis 27, 29–35 (2014).
Buchanan, E. L. et al. Expression and regulation of host restriction factor SAMHD1 during lentiviral infection of astrocytes and macrophages. In Conference on Retroviruses and Opportunistic Infections (Atlanta, GA, USA, 2013).
White, T. E. et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 (2013).
Cribier, A., Descours, B., Valadao, A. L., Laguette, N. & Benkirane, M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3, 1036–1043 (2013).
Hansen, E. C., Seamon, K. J., Cravens, S. L. & Stivers, J. T. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc Natl Acad Sci USA 111, E1843–1851 (2014).
Bloch, N. et al. HIV type 1 infection of plasmacytoid and myeloid dendritic cells is restricted by high levels of SAMHD1 and cannot be counteracted by Vpx. AIDS Res Hum Retroviruses 30, 195–203 (2014).
Zink, M. C. et al. High viral load in the cerebrospinal fluid and brain correlates with severity of simian immunodeficiency virus encephalitis. J Virol 73, 10480–10488 (1999).
Liu, J., Sisk, J., Gama, L., Clements, J. E. & Witwer, K. W. Tristetraprolin: expression and miRNA-mediated regulation during SIV infection of the central nervous system. Molecular Brain 6 (2013).
Sisk, J. M., Clements, J. E. & Witwer, K. W. miRNA Profiles of Monocyte-Lineage Cells Are Consistent with Complicated Roles in HIV-1 Restriction. Viruses 4, 1844–1864 (2012).
Flaherty, M. T., Hauer, D. A., Mankowski, J. L., Zink, M. C. & Clements, J. E. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J Virol 71, 5790–5798 (1997).
Christofidou-Solomidou, M., Pietrofesa, R., Arguiri, E., McAlexander, M. A. & Witwer, K. W. Dietary flaxseed modulates the miRNA profile in irradiated and non-irradiated murine lungs: A novel mechanism of tissue radioprotection by flaxseed. Cancer Biol Ther 15 (2014).
Acknowledgements
This work was supported in part by NIH grants R21 AI102659 (KWW), NIMH pilot grant P30 MH075673 (to KWW) and by the National Center for Research Resources and the Office of Research Infrastructure Programs (ORIP) and the National Institutes of Health through Grant Number P40 OD013117. DAE was supported by a Johns Hopkins University Provost’s Undergraduate Research Award for 2013, the Johns Hopkins Center for AIDS Research (1P30 AI094189) and a Dean’s Undergraduate Research Award for 2014. The authors are grateful to all members of the Molecular and Comparative Pathobiology Retrovirus Biology Laboratory for support and helpful discussions.
Author information
Authors and Affiliations
Contributions
K.W.W. conceived of, designed and managed the study. E.L.B., D.A.E., M.A.M., S.L.M., A.M. and K.W.W. performed experiments. E.L.B. and K.W.W. analyzed results and wrote the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Buchanan, E., Espinoza, D., McAlexander, M. et al. SAMHD1 transcript upregulation during SIV infection of the central nervous system does not associate with reduced viral load. Sci Rep 6, 22629 (2016). https://doi.org/10.1038/srep22629
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep22629
This article is cited by
-
Role of Intracellular Distribution of Feline and Bovine SAMHD1 Proteins in Lentiviral Restriction
Virologica Sinica (2021)
-
Increased SAMHD1 transcript expression correlates with interferon-related genes in HIV-1-infected patients
Medical Microbiology and Immunology (2019)
-
Immune Activation Influences SAMHD1 Expression and Vpx-mediated SAMHD1 Degradation during Chronic HIV-1 Infection
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.