Abstract
Meta-analyses of genome-wide association studies (meta-GWASs) and candidate gene studies have identified genetic variants associated with cardiovascular diseases, metabolic diseases and mood disorders. Although previous efforts were successful for individual disease conditions (single disease), limited information exists on shared genetic risk between these disorders. This article presents a detailed review and analysis of cardiometabolic diseases risk (CMD-R) genes that are also associated with mood disorders. First, we reviewed meta-GWASs published until January 2016, for the diseases ‘type 2 diabetes, coronary artery disease, hypertension’ and/or for the risk factors ‘blood pressure, obesity, plasma lipid levels, insulin and glucose related traits’. We then searched the literature for published associations of these CMD-R genes with mood disorders. We considered studies that reported a significant association of at least one of the CMD-R genes and ‘depression’ or ‘depressive disorder’ or ‘depressive symptoms’ or ‘bipolar disorder’ or ‘lithium treatment response in bipolar disorder’, or ‘serotonin reuptake inhibitors treatment response in major depression’. Our review revealed 24 potential pleiotropic genes that are likely to be shared between mood disorders and CMD-Rs. These genes include MTHFR, CACNA1D, CACNB2, GNAS, ADRB1, NCAN, REST, FTO, POMC, BDNF, CREB, ITIH4, LEP, GSK3B, SLC18A1, TLR4, PPP1R1B, APOE, CRY2, HTR1A, ADRA2A, TCF7L2, MTNR1B and IGF1. A pathway analysis of these genes revealed significant pathways: corticotrophin-releasing hormone signaling, AMPK signaling, cAMP-mediated or G-protein coupled receptor signaling, axonal guidance signaling, serotonin or dopamine receptors signaling, dopamine-DARPP32 feedback in cAMP signaling, circadian rhythm signaling and leptin signaling. Our review provides insights into the shared biological mechanisms of mood disorders and cardiometabolic diseases.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD), bipolar disorder (BPD), coronary artery diseases, type 2 diabetes and hypertension are amongst the major causes of disability, morbidity and mortality worldwide.1, 2 Although each of these conditions independently represent a major burden facing the health-care systems,1, 2, 3 their co-occurrence (co-morbidity) aggravates the situation and represents a challenge in psychosomatic medicine.4 Epidemiologically, MDD and BPD are bi-directionally associated with cardiometabolic diseases.5, 6 A similar pattern of association has been shown in the relationship between the pharmacological treatment of mood disorders and cardiometabolic diseases. For instance, the use of antidepressants and mood stabilizers is associated with an increased risk of cardiometabolic abnormalities7 and cardiac medications might increase the risk of mood disorders.8 One explanation for these relationships could be the presence of pleiotropic (common) genes and shared biological pathways that function as a hub to link the disorders. Potential common biological mechanisms underlying mood disorders and cardiometabolic disease comorbidity have been proposed, including altered circadian rhythms,9 abnormal hypothalamic–pituitary–adrenal axis (HPA axis) function,10 imbalanced neurotransmitters11 and inflammation.6 However, the molecular drivers of these commonly affected mechanisms remain poorly understood.
The genetics of mood disorders and cardiometabolic diseases
Major depression, bipolar disorder and cardiometabolic diseases are highly heritable and they are caused by a combination of genetic and environmental factors. Genetic factors contribute to 31-42% in MDD,12 59–85% in BPD,13, 14 30–60% in coronary artery diseases,15 26–69% in type 2 diabetes,16, 17 24–37% in blood pressure,18 40–70% in obesity19 and 58–66% in serum lipids level.20 Moreover, twin studies have revealed relatively modest genetic co-heritabilities (genetic correlations) between mood disorders and the different cardiometabolic abnormalities suggesting the influence of pleiotropic genes and shared biological pathways among them. For instance, the genetic correlation of depression with hypertension is estimated to be 19%, and between depression and heart disease is about 42%.21 The genetic correlation of depressive symptoms with plasma lipids level ranges from 10 to 31%,22 and 12% of the genetic component for depression is shared with obesity.23 Furthermore, gene–environment interactions can contribute to the cardiometabolic and mood disorders link. The interactions of genetic factors with stress, physical exercise, diet and lifestyle can influence the progression and pathogenesis of both cardiometabolic and mood disorders (Figure 1).24, 25, 26 These environmental factors might for example, modulate the expression of genes involved in the cardiometabolic pathways and a variety of pathways in the brain. Although it is at infancy stage, the ‘microbiome’ era has also revealed a range of complex interactions between environmental factors, genes and psychiatric disorders.27
Schematic model for the potential pleiotropic effects of a shared gene locus that is associated with mood disorders and cardiometabolic diseases.5, 6, 26, 70, 71, 75 The distal and proximal factors are obtained from the literature, and the World Health Organization (WHO) often uses the classification. Distal factors refer to those factors that require an intermediate factor to cause diseases, while proximal factors can directly cause diseases. The red bold lines represent the pleiotropic effect of a genetic locus on cardiometabolic diseases and associated risk factors, psychiatric morbidity, i.e.mood disorders and pharmacological treatment response in MDD and BPD. The bi-directional arrows indicate bidirectional epidemiological relationships between the cardiometabolic diseases and mood disorders. BPD, bipolar disorder; MDD, major depressive disorder.
In the last decade, substantial amounts of univariate (single disease) meta-analyses of genome-wide association studies (meta-GWASs) and candidate gene studies have been published. Indeed, the meta-GWASs and candidate gene studies have successfully identified a considerable list of candidate genes for major depressive disorder,28 bipolar disorder,29 coronary artery diseases,30 type 2 diabetes,31 hypertension,26 obesity,32 plasma lipids level,33 insulin and glucose traits31, 34 and blood pressure.26, 35
Despite the potential significance of studying pleiotropic genes and shared biological pathways, previous meta-GWAS and candidate gene studies were entirely focused on a single phenotype approach (single disease). A recent analysis of single-nucleotide polymorphisms (SNPs) and genes from the NHGRI GWAS catalog36 has showed as 16.9% of the genes and 4.6% of the SNPs have pleiotropic effects on complex diseases.37 Considering such evidence, we hypothesized that common genetic signatures and biological pathways mediate the mood disorders to cardiometabolic diseases relationship. In addition, these genes and their signalling pathways can influence the response to treatments in mood disorder patients (Figure 1). In this review, we systematically investigated the cardiometabolic diseases risk (CMD-R) genes that are possibly associated with mood disorders susceptibility, and with treatment response to MDD and BPD. We performed pathway and gene network analysis to provide additional insights in to the common pathways and biological mechanisms regulating mood disorders and the CMD-Rs. Understanding of these common pathways may provide new insights and novel ways for the diagnosis and treatment of comorbid cardiometabolic and mood disorders.
Materials and methods
Search strategy
Step 1: Identification of candidate genes for cardiometabolic diseases
We carried out a systematic search of candidate genes for the cardiometabolic diseases and/or associated risk factors. The National Human Genome Research Institute (NHGRI) GWAS catalogue,36 Westra et al.38 and Multiple Tissue Human Expression Resource (MuTHER)39 databases were used to identify the CMD-R genes. We reviewed meta-GWA study papers published until January 2016 for the diseases ‘type 2 diabetes’ or ‘coronary artery disease’ or ‘hypertension’ and (or) for the risk factors ‘blood pressure’ or ‘obesity or body mass index (BMI)’ or "plasma lipid levels (high-density lipoprotein, low-density lipoprotein, triglycerides, total cholesterol)’ or ‘insulin and glucose related traits (fasting glucose, fasting insulin, fasting proinsulin, insulin sensitivity, insulin resistance-HOMA-IR, beta cell function-HOMA-β and glycated haemoglobinA1C-HbA1C)’.
All GWAS significant SNPs (P<5 × 10−8) information (lead SNPs, reported genes, author(s), PubMed ID, date of publication, journal, discovery and replication sample sizes) was downloaded from the GWAS catalogue database. Additional information about the effect of the lead SNPs on nearby gene expression (cis-eQTLs) was collected from their respective publications. For the SNPs with no cis-eQTL information in their respective publications, we performed expression quantitative trait loci (cis-eQTL) analysis to verify the functional relationship between the reported genes and the lead SNPs using two publicly available databases: Westra et al.,38 and MuTHER.39 A CMD-R gene was considered as a candidate gene if, (1) at least one of the lead SNPs is located within or nearby to the gene; and (2) it is functionally relevant to influence at least one of the CMD-Rs as evidenced by gene expression analyses. We took the identified CMD-R genes forward for the second step literature review, as described below.
Step 2: Exploration of the role of cardiometabolic genes in mood disorders
In the second systematic review, we conducted a literature search in PubMed (MEDLINE database) for any genome wide association, candidate gene, or gene expression analysis study published in the fields of mood disorders and pharmacogenetics of mood disorders until January 2016. This step of the literature search was performed using SNIPPER tool (see web resources and tools). We considered studies that reported at least one of the CMD-R genes in ‘depression’ or ‘depressive disorder’ or ‘depressive symptoms’ or ‘MDD’ or ‘bipolar disorder’ or ‘mood disorder’ or ‘lithium treatment response’ or ‘Selective Serotonin Reuptake Inhibitors (SSRIs) treatment response’. A prior literature search implemented before the final review found that the majority of the genetic studies on treatment response to antidepressants and mood stabilizers were on lithium and SSRIs. As a result, the literature search on pharmacogenomics of mood disorders was limited to these predominant treatments.
Inclusion criteria
General inclusion criteria of genetic studies that involve individuals of all ages in both sexes was implemented. The pharmacogenomics studies were restricted to only lithium or SSRIs treatment response in mood disorders.
Exclusion criteria
Pharmacogenomics studies that used SSRIs or lithium for the treatment of psychosis, anxiety disorders, obsessive-compulsive disorder, post-traumatic stress disorder were excluded. We also excluded genetic studies that investigated drug-induced side effects of mood disorders.
Biological pathway and network analysis
The potential pleiotropic genes were further explored to identify the most enriched canonical pathways and visualize gene networks using QIAGEN's Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, CA, USA, www.qiagen.com/ingenuity). For the analysis, all the 24 potential pleiotropic genes were entered as input into the software. IPA compares the proportion of input genes mapping to a biological pathway to the reference genes in the ingenuity databases. The significance of the overrepresented canonical pathways were determined using the right-tailed Fisher’s exact test later adjusted for multiple testing using the Benjamini-Hochberg (BH) method.40 Significance levels were determined at BH adjusted P-value <0.01. A gene network that connects the input genes with MDD, BPD and the cardiometabolic disorders was also generated.
Web resources and tools
GWAS Catalogue: https://www.ebi.ac.uk/gwas/home
Westra et al. blood eQTL browser: http://genenetwork.nl/bloodeqtlbrowser/
MuTHER eQTL resource: http://www.muther.ac.uk/
SNIPPER tool v1.2: http://csg.sph.umich.edu/boehnke/snipper/
QIAGEN's Ingenuity Pathway Analysis: www.qiagen.com/ingenuity
Results
Characteristics of meta-GWA studies for the cardiometabolic disorders
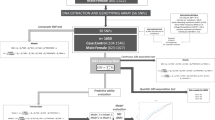
The literature searches in the GWAS catalogue yielded 153 meta-GWA studies for the CMD-Rs: 38 studies for type 2 diabetes, 17 studies for coronary artery disease, 15 studies for hypertension and blood pressure, 26 studies for obesity and BMI, 37 studies for lipids and 20 studies for glucose and insulin traits (Figure 2). As shown in Figure 2, the meta-GWA studies reported 1047 lead SNPs and 682 nearby genes. Of these, 123 genes were functionally relevant to the cardiometabolic diseases and associated risk factors, as confirmed by gene expression analysis (cis-eQTLs). These genes were reviewed for their association with mood disorders and pharmacogenetics of mood disorders. Twenty-four of the 123 CMD-R genes have been implicated in mood disorders; and we named these genes the Cardiometabolic Mood disorders hub (CMMDh) genes.
The flow chart shows the stages of literature search and evaluation of candidate pleiotropic genes for the CMD-Rs and mood disorders. CMD-R genes refers to the genes in which the CMD-R lead SNPs are located-in or nearby and their expression is influenced by the respective lead SNPs (cis-eQTL). CMD-R, Cardiometabolic Diseases and associated Risk factors; CMMDh, Cardiometabolic Mood Disorders hub genes; cis-eQTL, Cis (nearby) gene expression quantitative trait loci; GWAS, Genome Wide Assocation Study; Meta-GWA, meta-analysis of Genome Wide Association studies; MuTHER, Multiple Tissue Human Expression Resource; SNP, single nucleotide polymorphism.
Table 1 summarizes the 24 CMMDh genes and specific genetic variants across mood disorders and cardiometabolic diseases. These genes are MTHFR, CACNA1D, CACNB2, GNAS, ADRB1, NCAN, REST, FTO, POMC, BDNF, CREB, ITIH4, LEP, GSK3B, SLC18A1, TLR4, PPP1R1B, APOE, CRY2, HTR1A, ADRA2 A, TCF7L2, MTNR1B, and IGF1 (for further details see Table 1). These genes were over-represented in the following biological pathways: corticotrophin-releasing hormone signaling BDNF, CREB1, GNAS, POMC; AMPK signaling ADRA2A, ADRB1, CREB1, GNAS, LEP; cAMP-mediated and G-protein coupled receptor signaling ADRA2A, ADRB1, CREB1, GNAS, HTR1 A; axonal guidance signaling BDNF, GNAS, GSK3B, IGF1; serotonin and dopamine receptors signaling GNAS, HTR1A, SLC18A1, PPP1R1B; dopamine-DARPP32 feedback in cAMP PPP1R1B, CACNA1D, CREB1, GNAS; leptin signaling GNAS, LEP, POMC; and the circadian rhythm signaling CRY2, CREB1 (Table 2 and Figure 3).
The list of 24 CMMDh genes (left), genes enriched to the top canonical signaling pathways (middle) and the network of these genes with mood disorders and the CMD-Rs (right). In the right, it illustrates ingenuity IPA-generated network of the CMMDh genes with coronary artery diseases, hypertension, diabetes mellitus, obesity, depressive disorder and bipolar disorder. The coloured dotted lines highlights CMMDh genes that were related to bipolar disorder (orange) and depression (red). CMMDh, Cardiometabolic Mood Disorders hub genes; IPA, Ingenuity Pathway Analysis.
We also performed a gene network analysis of the CMMDh genes to the mood disorders and cardiometabolic diseases. On the basis of the network analysis, the CMMDh genes were centrally involved in the link between mood disorders and the cardiometabolic diseases. For instance, ADRB1 and ADRA2A genes linked the four most common cardiometabolic disorders (coronary diseases, hypertension, diabetes, obesity) with BPD and depressive disorder. The CACNB2 and CACNA1D genes have shown network with coronary diseases, hypertension, diabetes, BPD and depression. Similarly, the other CMMDh genes acted as a hub between at least one of the cardiometabolic disorders and BPD and/or depression (Figure 3).
Discussion
This, to the best of our knowledge, first cross-disorder review systematically evaluated candidate pleiotropic genes and biological pathways that are likely to be shared with mood disorders, cardiovascular diseases and metabolic disorders. We revealed 24 cardiovascular and metabolic disease genes implicated in depression, bipolar disorder or both. These genes belong to interrelated signaling pathways important in the hypotheses of both cardiometabolic diseases and mood disorders: corticotrophin-releasing hormone signaling, AMPK signaling, cAMP-mediated and G-protein-coupled receptor signaling, axonal guidance signaling, serotonin and dopamine receptors signaling, dopamine-DARPP32 feedback in cAMP signaling, leptin signaling and circadian rhythm signaling.
The corticotrophin-releasing hormone (CRH) signaling is one of the top canonical pathways that may underlie the link between CMD-Rs and mood disorders. This pathway comprises of CRH, CRH receptors (CRHR1, CRHR2), and other CRH-related peptides. It is the principal regulator of the HPA axis. There are consistent findings in the literature that support the role of the HPA axis dysregulation in mediating the risk of mood disorders and cardiovascular outcome.41 Our analysis found enriched CMMDh genes in the CRH signaling pathways (BDNF, CREB1, GNAS and POMC). Genetic variants of the genes for BDNF, CREB1, GNAS and POMC are associated with MDD,42, 43 BPD,44 obesity,32, 45 blood pressure and hypertension.35, 46 The genes belong to the group of stress responsive genes, and their activity could be modulated through the activation of the HPA-axis. In animal studies, the expression of BDNF47 and CREB1[ref. 48] genes were dysregulated by chronic stress. It is therefore possible that an interaction of BDNF, CREB1, GNAS, and POMC genes with exposure to chronic stress or traumatic life events increase the risk of cardiometabolic and mood disorders either simultaneously, or through mediating factors. The CRH signaling pathway is the principal regulator of stress responses.49 Following an exposure to stress, the hypothalamus releases the CRH, stimulating the secretion of adrenocorticotrophic hormone from the anterior pituitary gland. This in turn stimulates the adrenal gland to produce glucocorticoids (principally cortisol). Cortisol will then act on several organs including the brain through its receptors.49 In acute conditions, the production of cortisol helps the body to fight pathogens (stress) and alleviate inflammation. However, when stressors are long lasting (chronic) they can cause cortisol receptor resistance and failure of the HPA-axis negative-feedback mechanism. This increases the duration and chronicity of inflammation, and a failure to downregulate the inflammatory response. Ultimately, failure in the HPA-axis processes may cause dysfunction in the brain and the body, causing both somatic diseases and brain disorders. Stress can either originate from the external environment as chronic extrinsic stress (CES) or within the internal body system as chronic intrinsic stress (CIS). Both CES and CIS can influence the CRH pathway genes mainly through gene expression and DNA methylation mechanisms.50
In relation to stress, there are two possibilities to explain mood disorders to cardiometabolic diseases association. The first is that the human body system may consider mood disorders or CMD-Rs as CIS and then dysregulate the HPA-axis through the CRH signaling pathways. Given that mood disorders tend to have an earlier age of onset compared to most of the CMD-Rs,51 they might be the primary CIS to induce cardiometabolic outcomes through the CRH signaling mechanism. Another possibility is that CES and/or CIS interact with the CRH signaling genes to cause both CMD-Rs and mood disorders. In either of the conditions, the CRH signaling genes interacts with the stressors to cause a dysfunction in the HPA-axis.
The second main canonical pathway was the adenosine monophosphate-activated protein kinase (AMPK) signaling pathway. This pathway regulates the intercellular energy balance. It inhibits or induces ATP consuming and generating pathways as needed. The pathway is especially important for nerve cells, as they need more energy with small energy reserves.52 Abnormalities in the pathway can disturb normal brain functioning. In animal studies, Zhu et al., 2014 showed chronically stressed mice developed symptoms related to mood and metabolic abnormalities, such as significant weight gain, heightened anxiety, and depressive-like behavior. They also reported decreased levels of phosphorylated AMP-activated roteinkinase α (AMPKα), confirming the involvement of the AMPK pathway and its regulatory genes in metabolic disorders and depression.53 Recent studies also reported the activation of the AMPK pathway in rat hippocampus after ketamine treatment exerting rapid antidepressant effect.54 Major contributing CMMDh genes enriched in the AMPK pathway are ADRA2A, ADRB1, LEP, CREB1 and GNAS. Variations in one or more of these genes can influence the activity of the AMPK pathway, subsequently impairing energy homeostasis in the brain and possibly in other cells.52 This could later cause energy shortages for the brain and somatic cells. Since brain cells are the most vulnerable units that require substantial amount of energy supply, any energy shortage would severely affect first the brain. Symptoms of mood change such as depressive behavior could emerge during this process. Moreover, AMP activation, for instance during stress, could induce insulin resistance promoting metabolic syndrome, that is, obesity, diabetes and cardiovascular diseases.55, 56 Hence, it is very likely that inappropriate activity of the AMPK pathway can imbalance the energy needs of the cells and be a cause to mood disorders and cardiometabolic diseases.
Axonal guidance signaling was also among the top overrepresented canonical pathways. The pathway is essentially related to neuronal connections formed by the extension of axons, which migrate to reach their synaptic targets. Axon guidance is an important step in neural development. It allows growing axons to stretch and reach the next target axon to form the complex neuronal networks in the brain and throughout the body. The patterns of connection between nerves depend on the regulated action of guidance cues and their neuronal receptors that are themselves encoded by axonal guidance coding genes. Activation of specific signaling pathways can promote attraction or repulsion and affect the rate of axon extension. One important observation in the axonal guidance pathway is the role of calcium and voltage-dependent calcium channels. The pathway is regulated by the entrance of calcium through the plasma membrane and release from intracellular calcium store. Calcium has been implicated in controlling axon outgrowth.57 CMMDh genes overrepresented in the axonal guidance-signaling pathway include the BDNF, GNAS, GSK3B and IGF1 genes. Mutant axonal guidance genes followed by abnormal axon guidance and connectivity could cause a disorder primarily in the brain and subsequently to the peripheral organs.58
Other strong candidate mechanisms underlying mood disorders and cardiometabolic diseases are the serotonin and dopamine receptors signaling pathways. The serotonin pathway is mainly regulated by serotonin and its receptors known as 5-hydroxytryptamine receptors. Serotonin is a monoamine neurotransmitter synthesized in the central nervous system and its signaling modulates several physiological processes including regulation of appetite, mood and sleep, body temperature and metabolism. The SLC18A1, HTR1A and GNAS gene are among the CMMDh genes involved in the serotonin receptor-signaling pathway. The SLC18A1 gene encodes for the vesicular monoamine transporter that transports for monoamines. Its function is essential to the activity of the monoaminergic systems that have been implicated in several human neuropsychiatric disorders.59 The HTR1A gene encodes a receptor for serotonin, and it belongs to the 5-hydroxytryptamine receptor subfamily. Dysregulation of serotonergic neurotransmission has been suggested to contribute for the pathogenesis of mood disorders60, 61 and it is implicated in the action of selective serotonin reuptake inhibitors.62, 63, 64 Animal studies have consistently demonstrated the influence of the serotonin pathway on both mood disorders and cardiometabolic disorders. Ohta et al., 2011 have previously revealed as there is a converge in insulin and serotonin producing cells that can lead to metabolic diseases (diabetes) and mood disorders.65 The products of the insulin-producing cells (beta-islet cells) are involved to express the genes that synthesize serotonin, and serotonin also plays a role in the synthesis of insulin in the beta-islet cells.65
The dopamine receptors pathway, centrally regulated by dopamine, also appears to underlie the relationship between mood disorders and cardiometabolic diseases. Dopamine serves as a chemical messenger in the nervous system and its signaling has important roles in processes: emotion; positive reinforcement; motivation; movement; and in the periphery as a modulator of renal, cardiovascular and the endocrine systems.66 The SLC18A1 and GNAS genes are among the CMMDh genes that belong to this pathway. The dopamine-signaling pathway further induces the dopamine-DARPP32 Feedback in cAMP signaling. The central regulator of this pathway is the PPP1R1B gene that encodes a bifunctional signal transduction molecule called the dopamine and cAMP-regulated neuronal phosphoprotein (DARPP-32). Other CMMDh genes in the pathway include CACNA1D, CREB1, and GNAS. The CACNA1D gene encodes the alpha-1D subunit of the calcium channels that mediates the entry of calcium ions into excitable cells. Calcium channel proteins are involved in a variety of calcium-dependent processes, including hormone or neurotransmitter release, and gene expression.67
Overall, genes that encode for molecules involved in HPA-axis activity, circadian rhythm, inflammation, neurotransmission, metabolism and energy balance were found to have a central role to link mood disorders with cardiometabolic diseases. It is also worth noting the gene–environment interaction that might contribute to the diseases.
Implications of the review findings
Knowledge of genes and molecular pathways that are shared between mood disorders and cardiometabolic disorders have several important implications for future research and clinical practice. It is expected that increasing sample size, and consequently increasing power, will identify many more of the genes in the near future. Here we identify four implications of our findings.
First, the identification of shared molecular pathways implicated in disease susceptibility supports a growing evidence base for cross-diagnostic treatment paradigms. Shared molecular pathways could help to explain recent findings of reduced cardiovascular mortality,68 or improved diabetic control,69 in MDD patients treated with SSRIs. Second, further exploration of overlapping molecular pathophysiology has the potential to unveil novel targets for drug development, and may give clues for the re-purposing of existing medications.
Third, cardiometabolic disorders are associated with an increased risk of poor response to standard treatments in mood disorders.70, 71 Genetic profiling for cardiometabolic risk and stratified diagnosis of patients may help to classify treatment responders and treat them accordingly, thereby reducing the costs of ineffective exposure to medicines for the individuals and for the society. Early identification of at-risk individuals would also guide practitioner’s treatment recommendations, which may involve alternative somatic (for example, electroconvulsive therapy, repetitive Transcranial Magnetic Stimulation, ketamine) or specific psychological therapies as first- or second line treatments.
Fourth, studying the mechanisms of pleiotropic genes and shared pathways of mood disorders and somatic diseases could help untangle the clinical and genetic heterogeneity that characterizes these illnesses. It is possible that a ‘cardiometabolic’ endophenotype exists among mood disorders patients that may be identifiable through genetic profiling using polygenic scores or analysis of blood protein biomarkers. Preliminary evidence for such a phenotype, approximating the concept of ‘atypical depression’ characterized by increased appetite, weight gain and increased need for sleep, is emerging.72, 73 Working towards personalized care that allows for precise diagnostic, treatment and prevention strategies, research could then focus on genetically stratified patient cohorts instead of the very diverse patient pool currently diagnosed with MDD or BPD. There is a growing consensus that such stratification approaches have the potential to substantially improve the quality of mental health research and mental healthcare over the coming decades.74
Our review has limitations. Perhaps the most fundamental limitation was that almost all of the reviewed studies were performed in a univariate manner (single diseases approach). Essentially, multivariate models such as principal component analyses, multivariate mixed models and multivariate regression analyses are regarded as statistically powerful to perform cross-disorder analyses and identify pleiotropic genes. Unlike the multivariate approach, a univariate analysis investigates the association between a genetic variant and a single phenotype, aimed to identify genetic variants for individual diseases. Second, the review included studies that reported positively associated genes, and neither negative findings nor inconsistent evidences were assessed. We also found limited replication in some of the candidate genes, thereby demonstrating the necessity of future confirmatory studies. Third, only meta-GWAS were reviewed for the CMD-Rs and we implemented somewhat less stringent criteria for the genetic studies of mood disorders. GWAS for mood disorders have been less successful, mainly due to inadequate sample size and the phenotypic heterogeneity of the disorders. For this reason, the inclusion criteria for studies in these disorders was less strict. Hence, our review should be viewed as complementary to future mood disorders to cardiometabolic diseases gene investigation, providing an initial thorough summary of potential pleiotropic genes. Further population or case–control studies are necessary to confirm our proposed findings.
Conclusion
Our review revealed potential pleiotropic genes and biological pathways that are likely to be shared between mood disorders and cardiometabolic diseases. Although the review provides some insight into common mechanisms and the role of pleiotropic genes, in-depth understanding of how these genes (and possibly others) mediate the association between mood disorders and cardiometabolic diseases requires future comprehensive cross-disorder research in large-scale genetic studies. This will enable us to better understand why patients suffer from multiple diseases, and how multi-morbidities influence pharmacological treatment response to diseases.
References
Murray CJL, Barber RM, Foreman KJ, Ozgoren AA, Abd-Allah F, Abera SF et al. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990-2013: Quantifying the epidemiological transition. Lancet 2015; 386: 2145–2191.
Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet 2013; 382: 1575–1586.
Barnett K, Mercer SW, Norbury M, Watt G, Wyke S, Guthrie B . Epidemiology of multimorbidity and implications for health care, research, and medical education: A cross-sectional study. Lancet 2012; 380: 37–43.
Goldstein BI, Carnethon MR, Matthews KA, McIntyre RS, Miller GE, Raghuveer G et al. Major depressive disorder and bipolar disorder predispose youth to accelerated atherosclerosis and early cardiovascular disease: a scientific statement from the american heart association. Circulation 2015; 132: 965–986.
Golden SH, Lazo M, Carnethon M, Bertoni AG, Schreiner PJ, Roux AVD et al. Examining a bidirectional association between depressive symptoms and diabetes. JAMA 2008; 299: 2751–2759.
Kemp DE, Gao K, Chan PK, Ganocy SJ, Findling RL, Calabrese JR . Medical comorbidity in bipolar disorder: Relationship between illnesses of the endocrine/metabolic system and treatment outcome. Bipolar Disord 2010; 12: 404–413.
Correll CU, Detraux J, De Lepeleire J, De Hert M . Effects of antipsychotics, antidepressants and mood stabilizers on risk for physical diseases in people with schizophrenia, depression and bipolar disorder. World Psychiatry 2015; 14: 119–136.
Head GA . Impact of cardiac medications on mood In: Alvarenga M, Byrne D (eds) Handbook of Psychocardiology. Springer Singapore: Singapore, 2015; 1–14.
Zelinski EL, Deibel SH, McDonald RJ . The trouble with circadian clock dysfunction: multiple deleterious effects on the brain and body. Neuroscience and biobehavioral reviews 2014; 40: 80–101.
Rosmond R, Björntorp P . The hypothalamic-pituitary-adrenal axis activity as a predictor of cardiovascular disease, type 2 diabetes and stroke. J Int Med 2000; 247: 188–197.
Szczepanska-Sadowska E, Cudnoch-Jedrzejewska A, Ufnal M, Zera T . Brain and cardiovascular diseases: Common neurogenic background of cardiovascular, metabolic and inflammatory diseases. J Physiol Pharmacol 2010; 61: 509–521.
Sullivan PF, Neale MC, Kendler KS . Genetic epidemiology of major depression: Review and meta-analysis. Am J Psychiatry 2000; 157: 1552–1562.
Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373: 234–239.
McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A . The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry 2003; 60: 497–502.
Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U . Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 1994; 330: 1041–1046.
Almgren P, Lehtovirta M, Isomaa B, Sarelin L, Taskinen MR, Lyssenko V et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia 2011; 54: 2811–2819.
Poulsen P, Ohm Kyvik K, Vaag A, Beck-Nielsen H . Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance - A population-based twin study. Diabetologia 1999; 42: 139–145.
van Rijn MJE, Schut AFC, Aulchenko YS, Deinum J, Sayed-Tabatabaei FA, Yazdanpanah M et al. Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood-pressure-related genes. J Hypertens 2007; 25: 565–570.
Willyard C . Heritability: The family roots of obesity. Nature 2014; 508: S58–S60.
Knoblauch H, Busjahn A, Munter S, Nagy Z, Faulhaber H-D, Schuster H et al. Heritability analysis of lipids and three gene loci in twins link the macrophage scavenger receptor to HDL cholesterol concentrations. Arterioscler Thromb Vasc Biol 1997; 17: 2054–2060.
Scherrer JF, Xian H, Bucholz KK, Sa Eisen, Lyons MJ, Goldberg J et al. A twin study of depression symptoms, hypertension, and heart disease in middle-aged men. Psychosom Med 2003; 65: 548–557.
López-León S, Aulchenko YS, Tiemeier H, Oostra BA, van Duijn CM, Janssens ACJW . Shared genetic factors in the co-occurrence of symptoms of depression and cardiovascular risk factors. J Affect Dis 2010; 122: 247–252.
Afari N, Noonan C, Goldberg J, Roy-Byrne P, Schur E, Golnari G et al. Depression and obesity: Do shared genes explain the relationship? Depress Anxiety 2010; 27: 799–806.
Van Niel C, Pachter LM, Wade R Jr., Felitti VJ, Stein MT . Adverse events in children: predictors of adult physical and mental conditions. J Dev Behav Pediatr 2014; 35: 549–551.
Parnell LD, Blokker BA, Dashti HS, Nesbeth PD, Cooper BE, Ma Y et al. CardioGxE, a catalog of gene-environment interactions for cardiometabolic traits. BioData Min 2014; 7: 21.
Lopizzo N, Bocchio Chiavetto L, Cattane N, Plazzotta G, Tarazi FI, Pariante CM et al. Gene–environment interaction in major depression: focus on experience-dependent biological systems. Front Psychiatry 2015; 6: 68.
Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host/'s metabolism. Mol Psychiatry 2016; 21: 786–796.
Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 2016; 48: 1031–1036.
Psychiatric GCBDWG. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983.
Nikpay M, Goel A, Won H-H, Hall LM, Willenborg C, Kanoni S et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015; 47: 1121–1130.
Dupuis JJ, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010; 42: 105–116.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518: 197–206.
Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013; 45: 1274–1283.
Manning AK, Hivert M-F, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 2012; 44: 659–669.
Studies ICfBPG-WA Studies ICfBPG-WA Ehret GB Munroe PB Rice KM Bochud M Johnson AD et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011; 478: 103–109.
Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014; 42: D1001–D1006.
Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T et al. Abundant pleiotropy in human complex diseases and traits. J Hum GenetAm J Hum Genet 2011; 89: 607–618.
Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45: 1238–1243.
Grundberg E, Small KS, Hedman ÅK, Nica AC, Buil A, Keildson S et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 2012; 44: 1084–1089.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995; 57: 289–300.
Jokinen J, Nordstrom P . HPA axis hyperactivity and cardiovascular mortality in mood disorder inpatients. J Affect Disord 2009; 116: 88–92.
Zhang K, Yang C, Xu Y, Sun N, Yang H, Liu J et al. Genetic association of the interaction between the BDNF and GSK3B genes and major depressive disorder in a Chinese population. J Neural Transmisson 2010; 117: 393–401.
Zubenko GS, Hughes HB, Stiffler JS, Brechbiel A, Zubenko WN, Maher BS et al. Sequence variations in CREB1 cosegregate with depressive disorders in women. Mol Psychiatry 2003; 8: 611–618.
McDonald ML, MacMullen C, Liu DJ, Leal SM, Davis RL . Genetic association of cyclic AMP signaling genes with bipolar disorder. Transl Psychiatry 2012; 2: e169.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948.
Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet 2011; 43: 1005–1011.
Smith MA, Makino S, Kim SY, Kvetnansky R . Stress increases brain-derived neurotropic factor messenger ribonucleic acid in the hypothalamus and pituitary. Endocrinology 1995; 136: 3743–3750.
Grønli J, Bramham C, Murison R, Kanhema T, Fiske E, Bjorvatn B et al. Chronic mild stress inhibits BDNF protein expression and CREB activation in the dentate gyrus but not in the hippocampus proper. Pharmacol Biochem Behav 2006; 85: 842–849.
Hauger RL, Risbrough V, Brauns O, Dautzenberg FM . Corticotropin releasing factor (CRF) receptor signaling in the central nervous system: new molecular targets. CNS Neurol Disord Drug Targets 2006; 5: 453–479.
Mychasiuk R, Muhammad A, Kolb B . Chronic stress induces persistent changes in global DNA methylation and gene expression in the medial prefrontal cortex, orbitofrontal cortex, and hippocampus. Neuroscience 2016; 322: 489–499.
Katon W, Von Korff M, Ciechanowski P, Russo J, Lin E, Simon G et al. Behavioral and clinical factors associated with depression among individuals with diabetes. Diabetes Care 2004; 27: 914–920.
Ronnett GV, Aja S . AMP-activated protein kinase in the brain. Int J Obes (2005) 2008; 32 (Suppl 4): S42–S48.
Zhu S, Wang J, Zhang Y, Li V, Kong J, He J et al. Unpredictable chronic mild stress induces anxiety and depression-like behaviors and inactivates AMP-activated protein kinase in mice. Brain Res 2014; 1576: 81–90.
Xu SX, Zhou ZQ, Li XM, Ji MH, Zhang GF, Yang JJ . The activation of adenosine monophosphate-activated protein kinase in rat hippocampus contributes to the rapid antidepressant effect of ketamine. Behav Brain Res 2013; 253: 305–309.
Steinberg GR, Kemp BE . AMPK in health and disease. Physiol Rev 2009; 89: 1025–1078.
Lage R, Dieguez C, Vidal-Puig A, Lopez M . AMPK: a metabolic gauge regulating whole-body energy homeostasis. Trends Mol Med 2008; 14: 539–549.
Sutherland DJ, Pujic Z, Goodhill GJ . Calcium signaling in axon guidance. Trends Neurosci 2014; 37: 424–432.
Sasaki T, Oga T, Nakagaki K, Sakai K, Sumida K, Hoshino K et al. Developmental expression profiles of axon guidance signaling and the immune system in the marmoset cortex: potential molecular mechanisms of pruning of dendritic spines during primate synapse formation in late infancy and prepuberty (I). Biochem Biophys Res Commun 2014; 444: 302–306.
Peter D, Finn JP, Klisak I, Liu Y, Kojis T, Heinzmann C et al. Chromosomal localization of the human vesicular amine transporter genes. Genomics 1993; 18: 720–723.
Haenisch B, Linsel K, Brüss M, Gilsbach R, Propping P, Nöthen MM et al. Association of major depression with rare functional variants in norepinephrine transporter and serotonin1A receptor genes. Am J Med Genet B Neuropsychiatr Genet 2009; 150: 1013–1016.
Kishi T, Yoshimura R, Fukuo Y, Okochi T, Matsunaga S, Umene-Nakano W et al. The serotonin 1A receptor gene confer susceptibility to mood disorders: Results from an extended meta-analysis of patients with major depression and bipolar disorder. Eur Arch Psyhiatry Clin Neurosci 2013; 263: 105–118.
Baune BT, Hohoff C, Roehrs T, Deckert J, Arolt V, Domschke K . Serotonin receptor 1A -1019C/G variant: Impact on antidepressant pharmacoresponse in melancholic depression? Neurosci Lett 2008; 436: 111–115.
Serretti A, Artioli P, Lorenzi C, Pirovano A, Tubazio V, Zanardi R . The C(-1019)G polymorphism of the 5-HT1A gene promoter and antidepressant response in mood disorders: preliminary findings. Int J Neuropsychopharmacol 2004; 7: 453–460.
Arias B, Catalán R, Gastó C, Gutiérrez B, Fañanás L . Evidence for a combined genetic effect of the 5-HT(1A) receptor and serotonin transporter genes in the clinical outcome of major depressive patients treated with citalopram. J Pharmacol 2005; 19: 166–172.
Ohta Y, Kosaka Y, Kishimoto N, Wang J, Smith SB, Honig G et al. Convergence of the insulin and serotonin programs in the pancreatic beta-cell. Diabetes 2011; 60: 3208–3216.
Gordan R, Gwathmey JK, Xie LH . Autonomic and endocrine control of cardiovascular function. World J Cardiol 2015; 7: 204–214.
Pruitt K, Brown G, Tatusova T, Maglott D . The Reference Sequence (RefSeq) Database. National Center for Biotechnology Information: USA, 2012..
Acharya T, Acharya S, Tringali S, Huang J . Association of antidepressant and atypical antipsychotic use with cardiovascular events and mortality in a veteran population. Pharmacotherapy 2013; 33: 1053–1061.
Brieler JA, Lustman PJ, Scherrer JF, Salas J, Schneider FD . Antidepressant medication use and glycaemic control in co-morbid type 2 diabetes and depression. Fam Pract 2016; 33: 30–36.
Calkin CV, Ruzickova M, Uher R, Hajek T, Slaney CM, Garnham JS et al. Insulin resistance and outcome in bipolar disorder. Br J Psychiatry 2015; 206: 52–57.
Woo YS, Seo HJ, McIntyre RS, Bahk WM . Obesity and its potential effects on antidepressant treatment outcomes in patients with depressive disorders: a literature review. Int J Mol Sci 2016; 17: 1.
Milaneschi Y, Lamers F, Bot M, Drent ML, Penninx BW . Leptin dysregulation is specifically associated with major depression with atypical features: evidence for a mechanism connecting obesity and depression. Biol Psychiatry 2016 (in press).
Lamers F, Beekman AT, van Hemert AM, Schoevers RA, Penninx BW . Six-year longitudinal course and outcomes of subtypes of depression. Br J Psychiatry 2016; 208: 62–68.
Schumann G, Binder EB, Holte A, de Kloet ER, Oedegaard KJ, Robbins TW et al. Stratified medicine for mental disorders. Eur Neuropsychopharmacol 2014; 24: 5–50.
Calkin C, van de Velde C, Ruzickova M, Slaney C, Garnham J, Hajek T et al. Can body mass index help predict outcome in patients with bipolar disorder? Bipolar Disord 2009; 11: 650–656.
Lewis SJ, Lawlor DA, Davey Smith G, Araya R, Timpson N, Day INM et al. The thermolabile variant of MTHFR is associated with depression in the British Women's Heart and Health Study and a meta-analysis. Mol Psychiatry 2006; 11: 352–360.
Peerbooms OL, van Os J, Drukker M, Kenis G, Hoogveld L et alGroup MiP. Meta-analysis of MTHFR gene variants in schizophrenia, bipolar disorder and unipolar depressive disorder: evidence for a common genetic vulnerability? Brain Behav Immun 2011; 25: 1530–1543.
Lok A, Bockting CLH, Koeter MWJ, Snieder H, Assies J, Mocking RJT et al. Interaction between the MTHFR C677T polymorphism and traumatic childhood events predicts depression. Transl Psychiatry 2013; 3: e288.
Ament SA, Szelinger S, Glusman G, Ashworth J, Hou L, Akula N et al. Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci USA 2015; 112: 3576–3581.
McCarthy MJ, Le Roux MJ, Wei H, Beesley S, Kelsoe JR, Welsh DK . Calcium channel genes associated with bipolar disorder modulate lithium's amplification of circadian rhythms. Neuropharmacology 2016; 101: 439–448.
Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A et al. Genome-wide association study of blood pressure and hypertension. Nat Genet 2009; 41: 677–687.
Smoller JW . Identifi cation of risk loci with shared eff ects on fi ve major psychiatric disorders: A genome-wide analysis. Lancet 2013; 381: 1371–1379.
Klenke S, Siffert W . SNPs in genes encoding G proteins in pharmacogenetics. Pharmacogenomics 2011; 12: 633–654.
Zill P, Baghai TC, Engel R, Zwanzger P, Schüle C, Minov C et al. Beta-1-adrenergic receptor gene in major depression: influence on antidepressant treatment response. Am J Med Genet B Neuropsychiatr Genet 2003; 120B: 85–89.
Otsuki K, Uchida S, Wakabayashi Y, Matsubara T, Hobara T, Funato H et al. Aberrant REST-mediated transcriptional regulation in major depressive disorder. J Psychiatr Res 2010; 44: 378–384.
Goswami DB, May WL, Stockmeier CA, Austin MC . Transcriptional expression of serotonergic regulators in laser-captured microdissected dorsal raphe neurons of subjects with major depressive disorder: Sex-specific differences. J Neurochem 2010; 112: 397–409.
Hara K, Fujita H, Johnson TA, Yamauchi T, Yasuda K, Horikoshi M et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet 2014; 23: 239–246.
Kloiber S, Ripke S, Kohli MA, Reppermund S, Salyakina D, Uher R et al. Resistance to antidepressant treatment is associated with polymorphisms in the leptin gene, decreased leptin mRNA expression, and decreased leptin serum levels. Eur Neuropsychopharmacol 2013; 23: 653–662.
Eikelis N, Esler M, Barton D, Dawood T, Wiesner G, Lambert G . Reduced brain leptin in patients with major depressive disorder and in suicide victims. Mol Psychiatry 2006; 11: 800–801.
Haefner S, Baghai TC, Schule C, Eser D, Spraul M, Zill P et al. Impact of gene-gender effects of adrenergic polymorphisms on hypothalamic-pituitary-adrenal axis activity in depressed patients. Neuropsychobiology 2008; 58: 154–162.
Kato M, Serretti A, Nonen S, Takekita Y, Wakeno M, Azuma J et al. Genetic variants in combination with early partial improvement as a clinical utility predictor of treatment outcome in major depressive disorder: the result of two pooled RCTs. Transl Psychiatry 2015; 5: e513.
Wakeno M, Kato M, Okugawa G, Fukuda T, Hosoi Y, Takekita Y et al. The alpha 2A-adrenergic receptor gene polymorphism modifies antidepressant responses to milnacipran. J Clin Psychopharmacol 2008; 28: 518–524.
Perroud N, Aitchison KJ, Uher R, Smith R, Huezo-Diaz P, Marusic A et al. Genetic predictors of increase in suicidal ideation during antidepressant treatment in the GENDEP project. Neuropsychopharmacology 2009; 34: 2517–2528.
Cuffí ML, Artells R, Navarro A, Ciruela F, Carbonell L . Regulation of α2-adrenoceptor gene expression by chronic lithium treatment in rat brain. Methods Find Exp Clin Pharmacol 2010; 32: 721–725.
Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014; 46: 234–244.
Winham SJ, Cuellar-Barboza AB, Oliveros A, McElroy SL, Crow S, Colby C et al. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol Psychiatry 2014; 19: 1010–1016.
Zheng J-S, Arnett DK, Lee Y-C, Shen J, Parnell LD, Smith CE et al. Genome-wide contribution of genotype by environment interaction to variation of diabetes-related traits. PLoS ONE 2013; 8: e77442.
López-Figueroa AL, Norton CS, López-Figueroa MO, Armellini-Dodel D, Burke S, Akil H et al. Serotonin 5-HT1A, 5-HT1B, and 5-HT2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol Psychiatry 2004; 55: 225–233.
Carrard A, Salzmann A, Malafosse A, Karege F . Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J Affect Disord 2011; 132: 450–453.
Soria V, Martínez-Amorós E, Escaramís G, Valero J, Pérez-Egea R, García C et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 2010; 35: 1279–1289.
Geoffroy PA, Lajnef M, Bellivier F, Jamain S, Gard S, Kahn J-P et al. Genetic association study of circadian genes with seasonal pattern in bipolar disorders. Sci Rep 2015; 5: 10232.
Go MJ, Hwang J-Y, Kim YJ, Hee Oh J, Kim Y-J, Heon Kwak S et al. New susceptibility loci in MYL2, C12orf51 and OAS1 associated with 1-h plasma glucose as predisposing risk factors for type 2 diabetes in the Korean population. J Hum Genet 2013; 58: 362–365.
Chambers JC, Weihua Z, Zabaneh D, Sehmi J, Jain P, McCarthy MI et al. Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes 2009; 58: 2703–2708.
Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 2009; 41: 89–94.
Voight BF, Scott LJ, Steinthorsdottir V, Morris ADP, Dina C, Welch RP et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010; 42: 579–589.
Gałecka E, Szemraj J, Florkowski A, Gałecki P, Bieńkiewicz M, Karbownik-Lewińska M et al. Single nucleotide polymorphisms and mRNA expression for melatonin MT2 receptor in depression. Psychiatry Res 2011; 189: 472–474.
Kopczak A, Stalla GK, Uhr M, Lucae S, Hennings J, Ising M et al. IGF-I in major depression and antidepressant treatment response. Eur Neuropsychopharmacol 2015; 25: 864–872.
Mitschelen M, Yan H, Farley JA, Warrington JP, Han S, Hereñú CB et al. Long-term deficiency of circulating and hippocampal insulin-like growth factor I induces depressive behavior in adult mice: a potential model of geriatric depression. Neuroscience 2011; 185: 50–60.
Pereira ACP, McQuillin A, Puri V, Anjorin A, Bass N, Kandaswamy R et al. Genetic association and sequencing of the insulin-like growth factor 1 gene in bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet 2011; 156: 177–187.
Squassina A, Costa M, Congiu D, Manchia M, Angius A, Deiana V et al. Insulin-like growth factor 1 (IGF-1) expression is up-regulated in lymphoblastoid cell lines of lithium responsive bipolar disorder patients. Pharmacol Res 2013; 73: 1–7.
Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet 2013; 45: 501–512.
Samaan Z, Anand SS, Anand S, Zhang X, Desai D, Rivera M et al. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol Psychiatry 2013; 18: 1281–1286.
Rivera M, Cohen-Woods S, Kapur K, Breen G, Ng M, Butler A et al. Depressive disorder moderates the effect of the FTO gene on body mass index. Mol Psychiatry 2011; 17: 604–611.
Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet 2013; 22: 3597–3607.
Chang HS, Won ES, Lee HY, Ham BJ, Kim YG, Lee MS . The association of proopiomelanocortin polymorphisms with the risk of major depressive disorder and the response to antidepressants via interactions with stressful life events. J Neural Transm 2015; 122: 59–68.
Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang J-Y et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet 2014; 23: 5492–5504.
Finseth PI, Sønderby IE, Djurovic S, Agartz I, Malt UF, Melle I et al. Association analysis between suicidal behaviour and candidate genes of bipolar disorder and schizophrenia. J Affect Dis 2014; 163: 110–114.
Pandey GN, Rizavi HS, Ren X, Bhaumik R, Dwivedi Y . Toll-like receptors in the depressed and suicide brain. J Psychiatr Res 2014; 53: 62–68.
Hung YY, Kang HY, Huang KW, Huang TL . Association between toll-like receptors expression and major depressive disorder. Psychiatry Res 2014; 220: 283–286.
Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24.
Lohoff FW, Sander T, Ferraro TN, Dahl JP, Gallinat J, Berrettini WH . Confirmation of association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) gene and bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet 2005; 139B: 51–53.
Sarchiapone M, Carli V, Roy A, Iacoviello L, Cuomo C, Latella MC et al. Association of polymorphism (Val66Met) of brain-derived neurotrophic factor with suicide attempts in depressed patients. Neuropsychobiology 2008; 57: 139–145.
Kim B, Kim CY, Hong JP, Kim SY, Lee C, Joo YH et al. Brain-derived neurotrophic factor Val/Met polymorphism and bipolar disorder: Association of the met allele with suicidal behavior of bipolar patients. Neuropsychobiology 2008; 58: 97–103.
El-Hage W, Vourc'h P, Gaillard P, Léger J, Belzung C, Ibarguen-Vargas Y et al. The BDNF Val66Met polymorphism is associated with escitalopram response in depressed patients. Psychopharmacology 2014; 232: 575–581.
Pandey GN, Dwivedi Y, Rizavi HS, Ren X, Zhang H, Pavuluri MN . Brain-derived neurotrophic factor gene and protein expression in pediatric and adult depressed subjects. Prog Neuropsychopharmacol Biol Psychiatry 2010; 34: 645–651.
Hong W, Fan J, Yuan C, Zhang C, Hu Y, Peng D et al. Significantly decreased mRNA levels of BDNF and MEK1 genes in treatment-resistant depression. Neuroreport 2014; 25: 753–755.
Serretti A, Chiesa A, Calati R, Massat I, Linotte S, Kasper S et al. A preliminary investigation of the influence of CREB1 gene on treatment resistance in major depression. J Affect Dis 2011; 128: 56–63.
Murphy GM Jr., Sarginson JE, Ryan HS, O'Hara R, Schatzberg AF, Lazzeroni LC . BDNF and CREB1 genetic variants interact to affect antidepressant treatment outcomes in geriatric depression. Pharmacogenet Genomics 2013; 23: 301–313.
Li M, Luo X-J, Rietschel M, Lewis CM, Mattheisen M, Müller-Myhsok B et al. Allelic differences between Europeans and Chinese for CREB1 SNPs and their implications in gene expression regulation, hippocampal structure and function, and bipolar disorder susceptibility. Mol Psychiatry 2014; 19: 452–461.
Mamdani F, Alda M, Grof P, Young LT, Rouleau G, Turecki G . Lithium response and genetic variation in the CREB family of genes. Am J Med Genet B Neuropsychiatr Genet 2008; 147: 500–504.
Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 2009; 41: 47–55.
Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008; 40: 161–169.
Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 2009; 41: 56–65.
Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet 2011; 88: 372–381.
Schultz CC, Mühleisen TW, Nenadic I, Koch K, Wagner G, Schachtzabel C et al. Common variation in NCAN, a risk factor for bipolar disorder and schizophrenia, influences local cortical folding in schizophrenia. Psychol Med 2014; 44: 811–820.
Diniz BS, Talib LL, Joaquim HPG, de Paula VRJ, Gattaz WF, Forlenza OV . Platelet GSK3B activity in patients with late-life depression: marker of depressive episode severity and cognitive impairment? World J Biol Psychiatry 2011; 12: 216–222.
Saus E, Soria V, Escaramís G, Crespo JM, Valero J, Gutiérrez-Zotes A et al. A haplotype of glycogen synthase kinase 3β is associated with early onset of unipolar major depression. Genes Brain Behav 2010; 9: 799–807.
Lachman HM, Pedrosa E, Petruolo OA, Cockerham M, Papolos A, Novak T et al. Increase in GSK3beta gene copy number variation in bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 259–265.
Luykx JJ, Boks MP, Terwindt AP, Bakker S, Kahn RS, Ophoff RA . The involvement of GSK3beta in bipolar disorder: integrating evidence from multiple types of genetic studies. Eur Neuropsychopharmacol 2010; 20: 357–368.
Mitjans M, Arias B, Jiménez E, Goikolea JM, Sáiz PA, García-Portilla MP et al. Exploring genetic variability at PI, GSK3, HPA, and glutamatergic pathways in lithium response: association with IMPA2, INPP1, and GSK3B genes. J Clin Psychopharmacol 2015; 35: 600–604.
Iwahashi K, Nishizawa D, Narita S, Numajiri M, Murayama O, Yoshihara E et al. Haplotype analysis of GSK-3β gene polymorphisms in bipolar disorder lithium responders and nonresponders. Clin Neuropharmacol 2014; 37: 108–110.
Weissglas-Volkov D, Aguilar-Salinas CA, Nikkola E, Deere KA, Cruz-Bautista I, Arellano-Campos O et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J Med Genet 2013; 50: 298–308.
Ko A, Cantor RM, Weissglas-Volkov D, Nikkola E, Reddy PMVL, Sinsheimer JS et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat Commun 2014; 5: 3983.
Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet 2008; 40: 149–151.
Lohoff FW, Dahl JP, Ferraro TN, Arnold SE, Gallinat J, Sander T et al. Variations in the vesicular monoamine transporter 1 gene (VMAT1/SLC18A1) are associated with bipolar i disorder. Neuropsychopharmacology 2006; 31: 2739–2747.
Ishikawa M, Mizukami K, Iwakiri M, Asada T . Immunohistochemical and immunoblot analysis of Dopamine and cyclic AMP-regulated phosphoprotein, relative molecular mass 32,000 (DARPP-32) in the prefrontal cortex of subjects with schizophrenia and bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry 2007; 31: 1177–1181.
Kunii Y, Hyde TM, Ye T, Li C, Kolachana B, Dickinson D et al. Revisiting DARPP-32 in postmortem human brain: changes in schizophrenia and bipolar disorder and genetic associations with t-DARPP-32 expression. Mol Psychiatry 2014; 19: 192–199.
Coram MA, Duan Q, Hoffmann TJ, Thornton T, Knowles JW, Johnson NA et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am J Hum Genet 2013; 92: 904–916.
Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY et al. Large-scale genome-wide association studies in east Asians identify new genetic loci influencing metabolic traits. Nat Genet 2011; 43: 990–995.
Yen Y-C, Rebok GW, Gallo JJ, Yang M-J, Lung F-W, Shih C-H . ApoE4 allele is associated with late-life depression: a population-based study. Am J Geriatr Psychiatry 2007; 15: 858–868.
Acknowledgements
The University of Adelaide, Adelaide Graduate Centre supported this work. ATA was a recipient of a postgraduate study scholarship, Adelaide Scholarship International (ASI), from the University of Adelaide.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Amare, A., Schubert, K., Klingler-Hoffmann, M. et al. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. Transl Psychiatry 7, e1007 (2017). https://doi.org/10.1038/tp.2016.261
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2016.261
This article is cited by
-
Familial confounding of internalising symptoms and obesity in adolescents and young adults; a co-twin analysis
International Journal of Obesity (2024)
-
From Mendel to multi-omics: shifting paradigms
European Journal of Human Genetics (2024)
-
Association between type 2 diabetes and depressive symptoms after a 1-year follow-up in an older adult Mediterranean population
Journal of Endocrinological Investigation (2024)
-
Causal association between major depressive disorder and coronary heart disease: a two-sample bidirectional mendelian randomization study
BMC Medical Genomics (2023)
-
Impacts of gender and lifestyle on the association between depressive symptoms and cardiovascular disease risk in the UK Biobank
Scientific Reports (2023)