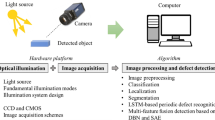
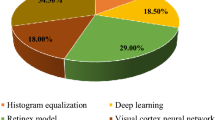
Abstract
Pathogenesis of many diseases is associated with changes in the collagen spatial structure. Traditionally, the 3D structure of collagen in biological tissues is analyzed using histochemistry, immunohistochemistry, magnetic resonance imaging, and X-radiography. At present, multiphoton microscopy (MPM) is commonly used to study the structure of biological tissues. MPM has a high spatial resolution comparable to histological analysis and can be used for direct visualization of collagen spatial structure. Because of a large volume of data accumulated due to the high spatial resolution of MPM, special analytical methods should be used for identification of informative features in the images and quantitative evaluation of relationship between these features and pathological processes resulting in the destruction of collagen structure. Here, we describe current approaches and achievements in the identification of informative features in the MPM images of collagen in biological tissues, as well as the development on this basis of algorithms for computer-aided classification of collagen structures using machine learning as a type of artificial intelligence methods.
Similar content being viewed by others
Abbreviations
- AGE:
-
advanced glycation end-product
- ANN:
-
artificial neural network
- AUC:
-
area under ROC curve
- CNN:
-
convolutional neural network
- CPA:
-
collagen proportional area
- DTI:
-
diffusion tensor imaging
- ECM:
-
extracellular matrix
- FLIM:
-
fluorescence lifetime imaging
- FOS:
-
first-order statistics
- GLCM:
-
gray-level co-occurrence matrix
- IR:
-
infrared
- MP:
-
metalloproteinase
- MPM:
-
multiphoton microscopy
- MRI:
-
magnetic resonance imaging
- PCA:
-
principal component analysis
- ReLU:
-
rectified linear unit
- ROC:
-
receiver operating characteristic
- ROS:
-
reactive oxygen species
- SHG:
-
second harmonic generation
- SIFT:
-
scale invariant feature transform
- SOS:
-
second-order statistics
- SVM:
-
support vector machine
- TCSPC:
-
time-correlated single photon counting
- TDS:
-
training dataset
- TPF:
-
two-photon fluorescence
- TPM:
-
two-photon microscopy
References
Andriotis, O. G., Chang, S. W., Vanleene, M., Howarth, P. H., Davies, D. E., Shefelbine, S. J., Buehler, M. J., and Thurner, P. J. (2015) Structure–mechanics relationships of collagen fibrils in the osteogenesis imperfecta mouse model, J. R. Soc. Interface, 12, 20150701.
Kapuler, O., Selskaya, B., Galeeva, A., and Kamilov, F. (2015) Metabolism of collagen fibers on the background of age–related changes, Vrach, No. 8, 64–69.
Lin, D., Chun, T. H., and Kang, L. (2016) Adipose extra–cellular matrix remodelling in obesity and insulin resist–ance, Biochem. Pharmacol., 119, 8–16.
http://www.medsest.ru/img/articles/vtorichnaya–struktura–v–vide–a–spirali–ili–v–skladchatoy–struktury.jpg
http://www.dcp–recovery.ru/p/blog–page_2.html
Simonenko, G. V., and Tuchin, V. V. (2007) Basic Knowledge about the Structure of Different Type Biological Tissues. Additional Knowledge from Human Anatomy [in Russian], Saratov State University, Saratov.
Gelse, K., Poeschl, E., and Aigner, T. (2003) Collagens − structure, function, and biosynthesis, Adv. Drug Deliv. Rev., 55, 1531–1546.
Mostaço–Guidolin, L. B., Ko, A. C.–T., Wang, F., Xiang, B., Hewko, M., Tian, G., Major, A., Shiomi, M., and Sowa, M. G. (2013) Collagen morphology and texture analysis: from statistics to classification, Sci. Rep., 3, 2190.
Aziz, J., Shezali, H., Radzi, Z., Yahya, N. A., Abu Kassim, N. H., Czernuszka, J., and Rahman, M. T. (2016) Molecular mechanisms of stress–responsive changes in col–lagen and elastin networks in skin, Skin Pharmacol. Physiol., 29, 190–203.
Said, G., Guilbert, M., Millerot–Serrurot, E., Van Gulick, L., Terryn, C., Garnotel, R., and Jeannesson, P. (2012) Impact of carbamylation and glycation of collagen type I on migration of HT1080 human fibrosarcoma cells, Int. J. Oncol., 40, 1797–1804.
Avery, N. C., and Bailey, A. J. (2006) The effects of the Maillard reaction on the physical properties and cell inter–actions of collagen, Pathol. Biol. (Paris), 54, 387–395.
Yuen, A., Laschinger, C., Talior, I., Leev, W., Chan, M., Birek, J., Young, E. W., Sivagurunathan, K., Won, E., Simmons, C. A., and McCulloch, C. A. (2010) Methylglyoxal–modified collagen promotes myofibroblast differentiation, Matrix Biol., 29, 537–548.
Kemeny, S. F., Figueroa, D. S., Andrews, A. M., Barbee, K. A., and Clyne, A. M. (2011) Glycated collagen alters endothelial cell actin alignment and nitric oxide release in response to fluid shear stress, J. Biomech., 44, 1927–1935.
Titov, V. N., Khokhlova, N. V., and Shiriaeva, I. K. (2013) Glucose, glycotoxins, and protein glycation products: the role in pathogenesis, Klin. Med. (Mosk.), 91, 15–24.
Tu, Y., and Quan, T. (2016) Oxidative stress and human skin connective tissue aging, Cosmetics, 3, 1–12.
Jablonska–Trypuc, A., Matejczyk, M., and Rosochacki, S. (2016) Matrix metalloproteinases (MMPS), the main extracellular matrix (ECM) enzymes in collagen degrada–tion, as a target for anticancer drugs, J. Enzyme. Inhib. Med. Chem., 31, 177–183.
Siwik, D. A., Pagano, P. J., and Colucci, W. S. (2001) Oxidative stress regulates collagen synthesis and matrix metalloproteinase activity in cardiac fibroblasts, Am. J. Physiol. Cell Physiol., 280, 53–60.
Rutkowski, J. M., Moya, M., Johannes, J., Goldman, J., and Swartza, M. A. (2006) Secondary lymphedema in the mouse tail: lymphatic hyperplasia, VEGF–C upregulation, and the protective role of MMP–9, Microvasc. Res., 72, 161–171.
Nikolaev, V. V., Kurochkina, A. S., Knyazkova, A. I., Vrazhnov, D. A., and Kistenev, Y. V. (2018) Study on lym–phedema disease using high resolution multiphoton microscopy and the FLIM technology, in Proc. XV Int. Conf. Students, Aspirants, Young Researchers “Prospects of Fundamental Science Development”.
Fang, M., Yuan, J., Peng, C., and Li, Y. (2014) Collagen as a double–edged sword in tumor progression, Tumour Biol., 35, 2871–2882.
Polasek, M., Yang, Y., Schuhle, D. T., Yaseen, M. A., Kim, Y. R., Sung, Y. S., Guimaraes, A. R., and Caravan, P. (2017) Molecular MR imaging of fibrosis in a mouse model of pancreatic cancer, Sci. Rep., 7, 1–10.
Zhang, L., Albon, J., Jones, H., Gouget, C. L. M., Ethier, C. R., Goh, J. C. H., and Girard, M. J. A. (2015) Collagen microstructural factors influencing optic nerve head bio–mechanics, Invest. Ophthalmol. Vis. Sci., 56, 2031–2042.
Pageon, H., Zucchi, H., Rousset, F., Monnier, V. M., and Asselineau, D. (2014) Skin aging by glycation: lessons from the reconstructed skin model, Clin. Chem. Lab. Med., 52, 169–174.
Gogola, A., Jan, N.–J., Brazile, B., Lam, P., Lathrop, K. L., Chan, K. C., and Sigal, I. A. (2018) Spatial patterns and age–related changes of the collagen crimp in the human cornea and sclera, Invest. Ophthalmol. Vis. Sci., 59, 2987–2998.
Navnita, S., Savita, S., Rithesh, K., and Shivaprasad, B. (2017) Evaluation of collagen degradation in gingival epithelium and connective tissue of chronic periodontitis patients with and without diabetes mellitus, Int. J. Sci. Res., 6, 33–35.
Tuchin, V. V. (2015) Tissue optics and photonics: light–tis–sue interaction, J. Biomed. Photonics Eng., 1, 98–135.
Topping, G., Malda, J., Dawson, R., and Upton, Z. (2006) Development and characterization of human skin equiva–lents and their potential application as a burn wound model, Primary Intention, 14, 14–21.
Oostendorp, C., Uijtdewilligen, P. J. E., Versteeg, E. M., Hafmans, T. G., van den Bogaard, E. H., de Jonge, P. K. J. D., Pirayesh, A., von den Hoff, J. W., Reichmann, E., Daamen, W. F., and van Kuppevelt, T. H. (2016) Visualization of newly synthesized collagen in vitro and in vivo, Sci. Rep., 6, 1–7.
Ross, K. A., Williams, R. M., Schnabel, L. V., Mohammed, H. O., Potter, H. G., Bradica, G., Castiglione, E., Pownder, S. L., Satchell, P. W., Saska, R. A., and Fortier, L. A. (2013) Comparison of three methods to quantify repair cartilage collagen orientation, Cartilage, 4, 111–120.
Jan, N.–J., Lathrop, K., and Sigal, I. A. (2017) Collagen architecture of the posterior pole: high–resolution widefield of view visualization and analysis using polarized light microscopy, Invest. Ophthalmol. Vis. Sci., 58, 735–744.
Fullerton, G. D., and Rahal, A. (2007) Collagen structure: the molecular source of the tendon magic angle effect, J. Magn. Reson. Imag., 25, 345–361.
Ghazanfari, S., Driessen–Mol, A., Strijkers, G. J., Baaijens, F. P. T., and Bouten, C. V. C. (2015) The evolu–tion of collagen fiber orientation in engineered cardiovas–cular tissues visualized by diffusion tensor imaging, PLoS One, 10, 1–15.
Johnson, G. A., Liu, C., Wei, H., Gibbs, E., Zhao, P., Wang, N., Cofer, G. P., and Zhang, Y. (2017) Susceptibility tensor imaging and tractography of collagen fibrils in the articular cartilage, Magn. Reson. Med., 78, 1683–1690.
Bashkatov, A. N., Genina, E. A., Kozintseva, M. D., Kochubei, V. I., Gorodkov, S. Y., and Tuchin, V. V. (2016) Optical properties of peritoneal biological tissues in the spectral range of 350–2500 nm, Opt. Spectrosc., 120, 1–8.
Yang, M. F., Tuchin, V. V., and Yaroslavsky, A. N. (2009) Principles of light–skin interactions, in Light–Based Therapies for Skin of Color (Baron, E., ed.) Springer, London, pp. 1–44.
Roth, S., and Freund, I. (1979) Second harmonic genera–tion in collagen, J. Chem. Phys., 70, 1637–1643.
Dumas, D., Werkmeister, E., Hupont, S., Huselstein, C., De Isla, N., Rousseau, M., Menu, P., and Mainard, D. (2014) Non–invasive second harmonic generation (SHG) in macroscopy (MacroSHG) as bio–diagnosis to image col–lagen network organization in extracellular matrix, Engineering, 6, 485–490.
Tuchin, V. V. (2006) Optical Clearing of Tissues and Blood, SPIE Press, Bellingham.
Tseng, J.–Y., Ghazaryan, A. A., Lo, W., Chen, Y.–F, Hovhannisyan, V., Chen, S.–J., Tan, H.–Y., and Dong, C.–Y. (2011) Multiphoton spectral microscopy for imaging and quantification of tissue glycation, Biomed. Opt. Express, 2, 218–230.
Huang, S., Heikal, A. A., and Webb, W. W. (2002) Two–photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein, Biophys. J., 82, 2811–2825.
Sdobnov, A., Darvin, M. E., Lademann, J., and Tuchin, V. (2017) A comparative study of ex vivo skin optical clearing using two–photon microscopy, J. Biophotonics, 10, 1115–1123.
Park, C. Y., Marando, C. M., Liao, J. A., Lee, J. K., Kwon, J., and Chuck, R. S. (2016) Details of the collagen and elastin architecture in the human limbal conjunctiva, Tenon’s capsule and sclera revealed by two–photon excited fluorescence microscopy, Invest. Ophthalmol. Vis. Sci., 57, 5602–5610.
Ambekar, R., Lau, T.–Y., Walsh, M., Bhargava, R., and Toussaint, K. C., Jr. (2012) Quantifying collagen structure in breast biopsies using second–harmonic generation imag–ing, Biomed. Opt. Express, 3, 2021–2035.
Golaraei, A., Kontenis, L., Cisek, R., Tokarz, D., Done, S. J., Wilson, B. C., and Barzda, V. (2016) Changes of collagen ultrastructure in breast cancer tissue determined by second–harmonic generation double Stokes–Mueller polarimetric microscopy, Biomed. Opt. Express, 7, 4054–4068.
Bogdanov, A. A., and Mazzanti, M. L. (2013) Fluorescent macromolecular sensors of enzymatic activity for in vivo imaging, Progr. Mol. Biol. Translat. Sci., 113, 349–387.
Vishwanath, V., Frank, K. E., Elmets, C. A., Dauchot, P. J., and Monnier, V. M. (1986) Glycation of skin collagen in type I diabetes mellitus correlation with long–term compli–cations, Diabetes, 35, 916–921.
Depeursinge, A., Fageot, J., and Al–Kadi, O. S. (2018) Fundamentals of texture processing for biomedical image analysis: a general definition and problem formulation, in Biomedical Texture Analysis: Fundamentals, Applications, Tools and Challenges (Depeursinge, A., Al–Kadi, O. S., and Ross Mitchell, J., eds.), Academic Press, pp. 1–27.
Castellano, G., Bonihla, L., Li, L. M., and Cendes, F. (2004) Texture analysis of medical images, Clin. Radiol., 59, 1061–1069.
Lowe, D. G. (1999) Object recognition from local scale–invariant features, Proc. 7th IEEE Int. Conf. Computer Vision, 2, 1150–1157.
Dalal, N., and Triggs, B. (2005) Histograms of oriented gradients for human detection, IEEE Computer Soc. Conf. Computer Vision Pattern Recognition, Vol. 1, pp. 886–893.
Donoho, D. L., and Huo, X. (1999) Combined image rep–resentation using edgelets and wavelets, Wavelet Applications Signal Image Processing VII, 3813, 468–477.
Huang, J., Kumar, R., Mitra, M., Zhu, W.–J., and Zabih, R. (1997) Image indexing using color correlograms, Proc. IEEE Computer Soc. Conf. Computer Vision and Pattern Recognition, pp. 762–768.
Guo, Z., Zhang, L., and Zhang, D. (2010) A completed modeling of local binary pattern operator for texture classi–fication, IEEE Transact. Image Process., 19, 1657–1663.
Simoncelli, E. P., and Freeman, W. T. (1995) The steerable pyramid: a flexible architecture for multi–scale derivative computation, Image Processing, Proc. Intern. Conf. IEEE, 3, 444–447.
Haralick, R. M., Sternberg, S. R., and Zhuang, X. (1987) Image analysis using mathematical morphology, IEEE Trans. Pattern Analysis Machine Intelligence, 4, 532–550.
Materka, A., and Strzelecki, M. (1998) Texture Analysis Methods–A Review, Technical University of Lodz, Institute of Electronics, COST B11 report, pp. 9–11.
Fletcher, N. D., and Evans, A. N. (2005) Texture segmen–tation using area morphology local granulometries, in Mathematical Morphology: 40Years On. Proc. 7th Int. Symp. on Mathematical Morphology (Ronse, C., Najman, L., and Decenciere, E., eds.) Springer, Dordrecht, pp. 367–376.
Bigun, J., Granlund, G. H., and Wiklund, J. (1991) Multidimensional orientation estimation with applications to texture analysis and optical flow, IEEE Trans. Pattern Analysis Machine Intelligence, 8, 775–790.
Kemao, Q. (2007) Two–dimensional windowed Fourier transform for fringe pattern analysis: principles, applica–tions and implementations, Opt. Lasers Eng., 45, 304–317.
Arivazhagan, S., and Ganesan, L. (2003) Texture classifica–tion using wavelet transform, Pattern Recogn. Lett., 24, 1513–1521.
Chang, T., and Kuo, C. C. J. (1993) Texture analysis and classification with tree–structured wavelet transform, IEEE Trans. Image Process., 2, 429–441.
Kistenev, Y. V., Shapovalov, A. V., Borisov, A. V., Vrazhnov, D. A., Nikolaev, V. V., and Nikiforova, O. Y. (2015) Wavelet based de–noising of breath air absorption spectra profiles for improved classification by principal component analy–sis, AIP Conf. Proc., 1688, 030010.
Khobragade, P., Fan, J., Rupcich, F., Crotty, D. J., and Schmidt, T. G. (2018) Application of fractal dimension for quantifying noise texture in computed tomography images, Med. Phys., 45, 3563–3573.
Depeursinge, A., Al–Kadi, O. S., and Mitchell, J. R. (2017) Biomedical Texture Analysis: Fundamentals, Tools and Challenges, Academic Press, London.
Sankar, D., and Thomas, T. (2010) Fractal features based on differential box counting method for the categorization of digital mammograms, J. Comput. Inform. Syst. Indust. Manag. Appl., 2, 11–19.
Bevk, M., and Kononenko, I. (2002) A statistical approach to texture description of medical images: a preliminary study, Proc. 15th IEEE Symp. Computer–Based Med. Systems, 64, 239–244.
Haralick, R. M., Shanmugam, K., and Dinstein, I. (1973) Textural features for image classification, IEEE Trans. Systems, Man, Cybernetics, SMC–3, 610–621.
Nanni, L., Brahnam, S., Ghidoni, S., Menegatti, E., and Barrier, T. (2013) Different approaches for extracting infor–mation from the co–occurrence matrix, PloS One, 8, e83554.
Farinella, G. M., Moltisanti, M., and Battiato, S. (2014) Classifying food images represented as bag of textons, 2014 IEEE Int. Conf. Image Processing (ICIP), 5212–5216.
Sahiner, B., Chan, H. P., Petrick, N., Wei, D., Helvie, M. A., Adler, D. D., and Goodsitt, M. M. (1996) Classification of mass and normal breast tissue: a convolution neural net–work classifier with spatial domain and texture images, IEEE Trans. Med. Imaging, 15, 598–610.
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., and Thrun, S. (2017) Dermatologist–level classification of skin cancer with deep neural networks, Nature, 542, 115–118.
Tivive, F. H. C., and Bouzerdoum, A. (2006) Texture clas–sification using convolutional neural networks, 2006 IEEE Region 10 Conf. (TENCON 2006), 1–4.
Anthimopoulos, M., Christodoulidis, S., Ebner, L., and Mouqiakakou, S. (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network, IEEE Trans. Med. Imaging, 35, 1207–1216.
Kistenev, Y. V., Borisov, A. V., Shapovalov, A. V., and Nikiforova, O. Y. (2015) Analysis of the component composi–tion of exhaled air using laser spectroscopy and canonical cor–relation analysis, Proc. SPIE 9680, 21st Int. Symp. Atmospheric and Ocean Optics: Atmospheric Physics, 9680, 96804C.
Postma, E., Herik, H. J., and Maaten, L. J. P. (2009) Dimensionality reduction: a comparative review, J. Mach. Learn. Res., 10, 1–41.
Nunes, M. A., Prangle, D., Sisson, S. A., and Blum, M. G. B. (2013) A comparative review of dimension reduction methods in approximate Bayesian computation, Stat. Sci., 28, 189–208.
Angryk, R., Martens, P., and Banda, J. M. (2012) Quantitative comparison of linear and non–linear dimen–sionality reduction techniques for solar image archives, Proc. 25th Int. FLAIRS Conf. (FLAIRS’12), pp. 376–381.
Kursa, M. B., and Rudnicki, W. R. (2010) Feature selection with the Boruta package, J. Stat. Soft., 36, 1–13.
Piatetsky–Shapiro, G., Matheus, C., and Frawley, W. (1992) Knowledge discovery in databases: an overview, AI Magazine, 13, 57–70.
Vrazhnov, D. A., Nikolaev, V. V., Shapovalov, A. V., and Sandykova, E. A. (2018) The kernel construction for the biomedical data classification using support vector machine, Proc. SPIE 10614, Int. Conf. Atomic and Molecular Pulsed Lasers XIII, 10614, 106141Y.
Arlot, S., and Celisse, A. (2010) A survey of cross–valida–tion procedures for model selection, Statist. Surv., 4, 40–79.
Hawkins, D. M. (2004) The problem of overfitting, J. Chem. Inf. Comp. Sci., 44, 1–12.
Boyd, S., and Vandenberghe, L. (2004) Convex Optimi–zation, Cambridge University Press, New York.
Hristu, R., Eftimie, L. G., Stanciu, S. G., Tranca, D. E., Paun, B., Sajin, M., and Stanciu, G. A. (2018) Quantitative second harmonic generation microscopy for the structural characterization of capsular collagen in thyroid neoplasms, Biomed. Opt. Express, 9, 3923–3936.
Liu, Z., Pouli, D., Sood, D., Sundarakrishnan, A., Hui Mingalone, C. K., Arendt, L. M., Alonzo, C., Quinn, K. P., Kuperwasser, C., Zeng, L., Schnelldorfer, T., Kaplan, D. L., and Georgakoudi, I. (2017) Automated quantifica–tion of three–dimensional organization of fiber–like struc–tures in biological tissues, Biomaterials, 116, 34–47.
Liang, L., Liu, M., and Sun, W. (2017) A deep learning approach to estimate chemically–treated collagenous tissue nonlinear anisotropic stress–strain responses from microscopy images, Acta Biomater., 63, 227–235.
Sacks, M. S. (2000) Biaxial mechanical evaluation of pla–nar biological materials, J. Elasticity, 61, 199–246.
Bekker, V., Zhelzov, A., and Cheslavsky, V. (2015) Visualization of fluorescence lifetime with the many–dimensional TCSPC–method: new prospects in biomedi–cine, Biofotonika, 5/53, 52–66.
Kistenev, Y. V., Nikolaev, V. V., Drozdova, A. K., Ilyasova, E. E., and Sandykova, E. A. (2018) Improvement of the multiphoton fluorescence microscopy images quality using digital filtration, Proc. SPIE 10614, Int. Conf. on Atomic and Molecular Pulsed Lasers XIII, 10614, 106141Q.
Mannan, R., Misra, V., Misra, S. P., Singh, P. A., and Dwivedi, M. (2014) A comparative evaluation of scoring systems for assessing necro–inflammatory activity and fibrosis in liver biopsies of patients with chronic viral hepa–titis, J. Clin. Diagn. Res., 8, FC08–FC12.
Tsipouras, M. G., Giannakeas, N., Tzallas, A. T., Tsianou, Z. E., Manousou, P., Hall, A., Tsoulos, I., and Tsianos, E. (2017) A methodology for automated CPA extraction using liver biopsy image analysis and machine learning tech–niques, Comput. Methods Programs Biomed., 140, 61–68.
Judd, N., Smith, J., Jain, M., Mukherjee, M., Icaza, M., Gallagher, R., Szeligowski, R., and Wu, B. (2018) A pilot study for distinguishing chromophobe renal cell carcinoma and oncocytoma using second harmonic generation imaging and convolutional neural network analysis of collagen fibrillar structure, Proc. SPIE 10489, Optical Biopsy XVI: Toward Real–Time Spectroscopic Imaging and Diagnosis, 10489, 1048919.
Kistenev, Y. V., Kuzmin, D. A., Vrazhnov, D. A., and Borisov, A. V. (2016) Classification of patients with broncho–pul–monary diseases based on analysis of absorption spectra of exhaled air samples with SVM and neural network algorithm application, Proc. SPIE 10035, 22nd Int. Symp. Atmospheric and Ocean Optics: Atmospheric Physics, 10035, 1003507.
Kistenev, Y. V., Borisov, A. V., Kuzmin, D. A., Penkova, O. V., Kostyukova, N. Y., and Karapuzikov, A. A. (2017) Exhaled air analysis using wideband wave number tuning range infrared laser photoacoustic spectroscopy, J. Biomed. Opt., 22, 17002.
Author information
Authors and Affiliations
Corresponding author
Additional information
Russian Text © Yu. V. Kistenev, D. A. Vrazhnov, V. V. Nikolaev, E. A. Sandykova, N. A. Krivova, 2019, published in Uspekhi Biologicheskoi Khimii, 2019, Vol. 59, pp. 219–252.
Rights and permissions
About this article
Cite this article
Kistenev, Y.V., Vrazhnov, D.A., Nikolaev, V.V. et al. Analysis of Collagen Spatial Structure Using Multiphoton Microscopy and Machine Learning Methods. Biochemistry Moscow 84 (Suppl 1), 108–123 (2019). https://doi.org/10.1134/S0006297919140074
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297919140074