Abstract
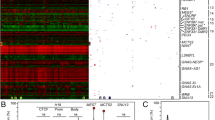
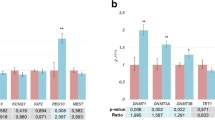
A complex network of epigenetic factors participates in regulating the monoallelic expression of a small subset of genes (˜1%) in the human genome. This phenomenon goes under the definition of genomic imprinting, a parent-of-origin effect that, when altered during early embryogenesis, may influence fetal development into adulthood. Pertubations in genomic imprinting have been associated with placental and fetal growth restrictions. We analyzed the differential DNA methylation of all known imprinted genes on 10 appropriate-for-gestational-age, clinically normal, placentas and 7 severe intrauterine growth-restricted placentas. Samples were pooled according to the diagnosis and analyzed by methylated DNA immunoprecipitation (MeDIP) on a tiling microarray platform. The distribution of the differentially methylated regions (DMRs) identified in growth-restricted placentas showed a slight tendency toward hypermethylation. Imprinted genes not expressed in placenta showed a unique DMR profile with the fewest hyper- and hypomethylated DMRs. Promoter and CpG island DMRs were sporadic and randomly distributed. The vast majority of DMR identified (˜99%) were mapped in introns, showing no common sequence features. Also, by using the more advanced array data mining softwares, no significant patterns emerged. In contrast, differential methylation showed a highly significant correlation with gene length. Overall these data suggest that differential methylation changes in growth-restricted placentas occur throughout the genomic regions, encompassing genes actively expressed in the placenta. These findings warrant caution in interpreting the significance of genes carrying clustered DMRs because the distribution of DMRs in a gene may be attributed as a function of its length rather than as a specific biological role.
Similar content being viewed by others
References
Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat Methods. 2008;5(4):307–309.
Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14(2):188–195.
Coan PM, Burton GJ, Ferguson-Smith AC. Imprinted genes in the placenta-a review. Placenta. 2005;26(suppl A):S10–S20.
Kawahara M, Morita S, Takahashi N, Kono T. Defining contributions of paternally methylated imprinted genes at the Igf2-H19 and Dlk1-Gtl2 domains to mouse placentation by transcriptomic analysis. J Biol Chem. 2009;284(26):17751–17765.
Tycko B. Imprinted genes in placental growth and obstetric disorders. Cytogenet Genome Res. 2006;113(1–4):271–278.
Walter J, Paulsen M. Imprinting and disease. Semin Cell Dev Biol. 2003;14(1):101–110.
Monk D, Bentley L, Hitchins M, et al. Chromosome 7p disruptions in Silver Russell syndrome: delineating an imprinted candidate gene region. Hum Genet. Oct 2002;111(4–5):376–387.
Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15(Spec No 2):R138–R150.
Susser E, Hoek HW, Brown A. Neurodevelopmental disorders after prenatal famine: the story of the Dutch Famine Study. Am J Epidemiol. 1998;147(3):213–216.
Svensson AC, Pawitan Y, Cnattingius S, Reilly M, Lichtenstein P. Familial aggregation of small-for-gestational-age births: the importance of fetal genetic effects. Am J Obstet Gynecol. 2006;194(2):475–479.
Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14(1):3–12.
Diplas AI, Lambertini L, Lee MJ, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4(4):235–240.
Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186.
Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69–73.
Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637.
Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2(6):e137.
Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 2007;39(8):945–949.
Newman JR, Ghaemmaghami S, Ihmels J, et al. Single-cell proteo-mic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846.
Zaitoun I, Downs KM, Rosa GJ, Khatib H. Upregulation of imprinted genes in mice: an insight into the intensity of gene expression and the evolution of genomic imprinting. Epigenetics. 2010;5(2):149–158.
Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476.
Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigen-etically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102(2):419–427.
Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta Proc Natl Acad Sci U S A. 2006;103(14): 5478–5483.
Chen J, Germer S, Higuchi R, Berkowitz G, Godbold J, Wetmur JG. Kinetic polymerase chain reaction on pooled DNA: a high-throughput, high-efficiency alternative in genetic epidemiological studies. Cancer Epidemiol Biomarkers Prev. 2002;11(1):131–136.
Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci U S A. 2005;102(12):4252–4257.
Macgregor S. Most pooling variation in array-based DNA pooling is attributable to array error rather than pool construction error. Eur J Hum Genet. 2007;15(4):501–504.
Zhang W, Carriquiry A, Nettleton D, Dekkers JC. Pooling mRNA in microarray experiments and its effect on power. Bioinfor-matics. 2007;23(10):1217–1224.
Gieni RS, Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem Cell Biol. 2009;87(5):711–746.
Guo L, Choufani S, Ferreira J, et al. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol. 2008;320(1):79–91.
Shen HM, Nakamura A, Sugimoto J, et al. Tissue specificity of methylation and expression of human genes coding for neuropeptides and their receptors, and of a human endogenous retrovirus K family. J Hum Genet. 2006;51(5):440–450.
Lewis A, Reik W. How imprinting centres work. Cytogenet Genome Res. 2006;113(1–4):81–89.
Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus Nature. 2000;405(6785): 486–489.
Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta Epigenetics. 2008;3(5): 261–269.
Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lambertini, L., Lee, TL., Chan, WY. et al. Differential Methylation of Imprinted Genes in Growth-Restricted Placentas. Reprod. Sci. 18, 1111–1117 (2011). https://doi.org/10.1177/1933719111404611
Published:
Issue Date:
DOI: https://doi.org/10.1177/1933719111404611