
Profiles of differentially expressed long noncoding RNAs and messenger RNAs in the myocardium of septic mice
Introduction
Sepsis, which is now defined as life-threatening organ dysfunction caused by the deregulated host response to infection, is currently the leading cause of mortality in the intensive care unit (ICU) (1,2). In the United States, admissions for sepsis have exceeded those for myocardial infarction and stroke (3). According to a recent epidemiological study, there are approximately 5 million patients with sepsis in China each year, and the mortality rate is greater than 30% (4). Therefore, although enormous improvements have been made in treating sepsis, such as anti-infection strategy and extracorporeal life support technology, the number of deaths is still rising steadily (5), and sepsis remains the major challenge for intensive care physicians (6,7). Numerous evidence-based clinical studies have shown that the fundamental cause of the high mortality rate in patients with sepsis is the dysfunction and failure of essential organs, such as the heart and lung (8,9).
Septic myocardial dysfunction (SMD) as a critical component of sepsis-induced multiple organ dysfunction is closely associated with adverse outcomes and high mortality (10). For example, in contrast to 20% mortality for septic patients without cardiac involvement, the mortality rate of patients with SMD ranges from 70% to 90% (11). Thus, effectively suppressing multiple organ dysfunction, particularly SMD, is crucial to improve the prognosis of septic patients.
Long noncoding RNAs (lncRNAs), characterized by a transcript of more than 200 nucleotides without the capability to translate into proteins, have long been considered as part of transcriptional noise (12). However, increasing evidence has demonstrated that lncRNAs are novel regulators of chromatin remodeling, transcriptional, and post-transcriptional gene regulation (12). Recently, the potential role of lncRNAs in the pathogenesis of SMD has attracted some attention. Besides, several lncRNAs have been reported to correlate with SMD (13-17). In animal studies, the overexpression of lncRNA CRNDE played a protective role against sepsis-induced cardiomyocyte apoptosis and oxidative damage by modulating the microRNA-29a/SIRT1 axis (16); similarly, sepsis-induced cardiomyocyte apoptosis was significantly increased when lncRNA Pvt1 was silenced (17). In human studies, plasma levels of at least four lncRNAs in septic patients were substantially different from those in healthy volunteers, suggesting that circulating lncRNAs are a novel noninvasive diagnostic biomarker of SMD (18). However, the precise role of lncRNAs in SMD pathogenesis is still unclear, especially the expression profiles of lncRNAs and messenger RNAs (mRNAs) and the interacting pathways between lncRNAs and mRNAs.
In this study, microarray technology was used to analyze the expression profiles of lncRNAs and mRNAs in septic mice’s hearts, thus evaluating the feasibility of lncRNAs as potential treatment targets for SMD.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3830).
Methods
Ethical statement
Experiments were performed under a project license (NO.: SHDSYY-2018-1524) granted by the Animal Ethics Committee of Shanghai Tenth People’s Hospital, Tongji University, in compliance with the guidelines described in the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Study design
Specific pathogen-free (SPF) male C57BL/6 mice (21–25 g) were purchased from Shanghai Laboratory Animal Co. Ltd. (Shanghai, China) and housed in the animal room (SPF class) of Shanghai Tenth People’s Hospital. All mice were labeled and divided into experimental and control groups using a computer-based randomized digital method. Mice in the experimental group were modeled to sepsis, and only sham operation was conducted in the control group. Samples were taken to verify the expression profiles of lncRNAs and mRNAs in the myocardial tissues 24 h after the procedure.
Sepsis model
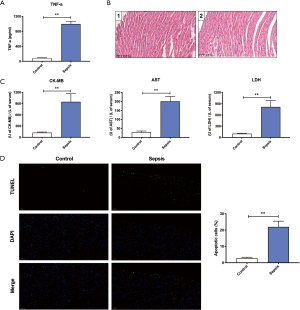
Cecal ligation and puncture (CLP) was performed as previously described to induce sepsis (19). The mouse was anesthetized with pentobarbital (75 mg/kg) intraperitoneally. The abdomen’s lower quadrant was trimmed using an electric shaver and disinfected with alcohol cotton balls three times. In the midline of the abdominal skin, a longitudinal incision was made with a scalpel. Small scissors were used to lengthen the incision to facilitate access to the peritoneal cavity. A 1-cm incision was made along with the linea alba to locate and exteriorize the cecum. Black-braided silk nonabsorbable suture (4-0) was used to ligate the cecum to acquire the middle grade of sepsis according to anatomical locations. The cecum was then perforated with a 21-G needle by a one-way through-and-through puncture between the ligation and the end of the cecum from mesenteric to the anti-mesenteric direction. Finally, the cecum was put back into the abdominal cavity, and the peritoneum, fasciae, abdominal musculature, and skin were closed in proper order. Postoperatively, animals were recovered by injecting pre-heated normal saline (37 °C; 5 mL/100 g) subcutaneously. In sham-operated mice (the control group), the operation was done identically, except that the cecum was not ligated and punctured. All animals were sacrificed humanely by an overdose of pentobarbital 24 h after surgery, and hearts were collected promptly for further studies. To verify that SMD was induced successfully by CLP, histological analysis, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) immunostaining, and Western blot analysis were performed in heart tissue samples. Plasma levels of myocardial enzymes and tumor necrosis factor-α (TNF-α) were also determined (Figure 1 and Figure S1).
Enzyme-linked immunosorbent assay (ELISA)
The concentration of serum TNF-α was determined by an ELISA kit (70-EK282/3-96, Multi Sciences Biotech Co. Ltd., Hangzhou, China) following the instruction manual. A microplate reader (Synergy 4 Hybrid Microplate Reader, BioTek, Vermont, USA) was used to measure the optical density spectrophotometrically at 450 and 630 nm.
Histological analysis of heart tissue
Mice were perfused transcardially with 0.9% normal saline. Fresh heart tissue samples were then collected, fixed with 4% paraformaldehyde, and embedded in paraffin. After deparaffinization and dehydration, transverse sections (6 µm) were stained with hematoxylin-eosin and analyzed under a light microscope.
TUNEL immunostaining
For TUNEL staining, it was carried with a One Step TUNEL Apoptosis Assay Kit (C1086, Beyotime Biotechnology, Shanghai, China) in accordance with the manufacturer’s instructions, and the slides were finally viewed under a fluorescence microscope (IX71, Olympus, Tokyo, Japan). The ratio of cell apoptosis was calculated as the percentage of apoptotic nuclei/the total number of nuclei in 10 randomly selected areas.
Biochemical analysis
Plasma levels of creatine aspartate aminotransferase (AST), lactate dehydrogenase (LDH), and creatine kinase-MB (CK-MB) were determined by a VITROS 5600 automated biochemical analyzer (Ortho Clinical Diagnostics, New York, USA).
RNA extraction
The total RNA of mouse hearts was extracted in TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to the instruction and operation manual. Briefly, isolated hearts were ground in 1 mL TRIzol reagent, and 200 µL chloroform was added to the myocardial tissue’s homogenate to extract RNA. The supernatant (600 µL) was transferred to a new Eppendorf (EP) tube after centrifugation at 12,000 rpm for 15 min at 4 °C, and the same volume of isopropanol was added. The mixed solution was vortexed sharply for 15 s, kept at room temperature for 10 min, and then centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatant was removed, and the RNA pellet was washed with 75% alcohol two times, dried on an operating platform at room temperature, and dissolved in diethyl pyrocarbonate treated water. A NanoDrop ND-1000 was used to determine RNA quality and quantity, and the integrity of RNA was evaluated by standard denaturing agarose gel electrophoresis (Figure S2).
Microarray analysis
The Arraystar Mouse LncRNA Microarray V3.0 (Arraystar, Rockville, MD, USA) was applied to profile mouse lncRNAs and protein-coding transcripts. Sample marking and array hybridization were carried out in accordance with the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology, USA) with a slight adjustment. In short, after removing rRNAs, the mRNA was purified from total RNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Each specimen was then amplified and transcribed into fluorescent complementary RNA (cRNA) along the transcripts’ full length without 3' bias using a randomized priming method (Arraystar Flash RNA Labeling Kit, Arraystar). The labeled cRNAs were purified by RNeasy Mini Kit (Qiagen, Hilden, Germany). The density and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were determined by NanoDrop ND-1000. Each labeled cRNA (1 µg) was fragmented by adding 5 µL 10× blocking agent and 1 µL of 25× fragmentation buffer, then warmed up the mixed solution at 60 °C for 30 min, 25 µL 2× GE hybridization buffer was added to dilute the labeled cRNA. The hybridization solution (50 µL) was dispensed with the gasket slide and assembled on the lncRNA expression microarray slide. The slides were incubated at 65 °C for 17 h in an Agilent hybridization oven. In the end, the hybridized arrays were washed, fixed, and scanned by the Agilent DNA Microarray Scanner (part number G2505C).
Quantitative real-time polymerase chain reaction (qRT-PCR)
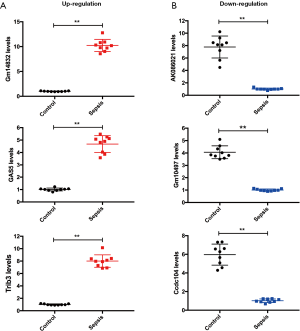
To identify microarray data’s replicability by qRT-PCR, a computer-based randomized digital method was chosen to randomly select six lncRNAs from the differentially expressed lncRNAs. In brief, the top 80 up-regulated or down-regulated lncRNAs were numbered from 1 to 80, respectively. The starting point and the order of sampling were randomly determined to generate a random number table by a computer. Finally, three up-regulated lncRNAs (Gm14832, GAS5, and Trib3) and three down-regulated lncRNAs (AK086021, Gm10497, and Ccdc104) were selected respectively from the top 80 up-regulated or down-regulated lncRNAs by extracting three random numbers in turn. The cDNA synthesis kit (RR036A, Takara Bio Inc., Shiga, Japan) was used to synthesize cDNA from RNA according to the kit’s instructions. The gene expression of cDNA was detected by the SYBR® Premix Ex Taq™ II (RR820A, Takara Bio Inc., Shiga, Japan) with an automated PCR instrument (7500 system). The reaction conditions were the following: incubation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s, and finally annealing and extension at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. The specific gene primer sequences are listed in Table S1. β-actin was selected as an internal control.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
GO and KEGG pathway analyses were performed to clarify the roles of all differentially expressed mRNAs as previously described (20). GO analysis was conducted to identify the gene and gene product enrichment, and therefore elucidate the biological roles of all aberrantly expressed mRNAs (http://www.geneontology.org). The GO covered three domains: biological processes (BP), cellular components (CC), and molecular functions (MF). Based on pathway analysis, KEGG pathway analysis (http://www.genome.jp/kegg/pathway.html) is designed for the functional mapping of genes. The thresholds to define markedly enriched GO terms/pathways were P≤0.05 and fractional disappearance rate ≤0.05.
Construction of lncRNA-mRNA co-expression network
The co-expression network was constructed to identify any potential interactions between the differentially expressed lncRNAs and mRNAs according to correlation analysis. The lncRNA-mRNA co-expression network’s rationale is the standardized signal intensities of specific lncRNA and mRNA expression levels. Cytoscape software (version 2.8.3, the Cytoscape Consortium, San Diego, CA, USA) was used to construct the lncRNA-mRNA co-expression network. The Pearson correlation coefficients (PCCs) were applied here to devise the lncRNA-mRNA co-expression network, PCCs ≥0.9 were chosen to build the network.
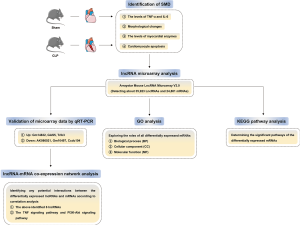
The primary method steps of this study are summarized in Figure 2.
Statistical analysis
All results were calculated and analyzed using SPSS 21.0 statistical software package (SPSS Inc., USA). The grouping was performed randomly; all data were presented as mean ± standard deviation (SD). Fold change (FC) and independent sample t-test were used to analyze the statistically significant difference in microarray results. The cut-off values for differentially expressed lncRNAs and mRNAs were FC ≥2 and P≤0.05. The false discovery rate (FDR) was determined to acquire the corrected P value, as described by Benjamini and Hochberg (21,22).
Results
Myocardial injury by CLP-induced sepsis
In the experimental group, CLP caused a significant increase in serum TNF-α concentration, indicating the occurrence of sepsis (Figure 1A). Myocardial injury due to CLP was verified by abnormal morphological changes (Figure 1B) and markedly increased myocardial enzymes (Figure 1C) in the experimental group. In the myocardium, the myocardial injury was also proved by significantly increased expression of apoptosis-related proteins B-cell lymphoma-2 (Bcl-2) and Bcl-2-associated X protein (Bax), and positive TUNEL staining. The ratio of apoptosis-positive cells in myocardial tissue was distinctly increased in CLP mice than in sham-operated mice (Figure 1D). In the septic mice, the expression level of Bcl-2 was notably reduced, the level of Bax was considerably increased, and the ratio of Bcl-2 to Bax was markedly decreased (Figure S1).
Expression profiles of lncRNAs and mRNAs in septic myocardium
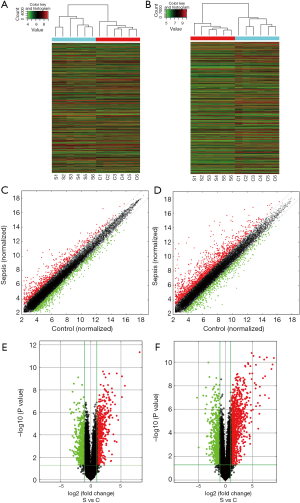
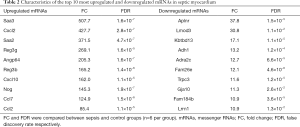
Six paired myocardial samples (sepsis and control groups, respectively) were collected for microarray analysis. The hierarchical clustering technique was used to illustrate the differential lncRNAs and mRNAs expression profiles in sepsis and control groups (Figure 3A,B). Subsequently, the heterogeneity of lncRNAs and mRNAs expression in these two groups was shown in the scatter plots (Figure 3C,D). With the criteria as FC ≥2 and P≤0.05, volcano plots were used to further identify the association between the FCs and the statistically significant difference of differentially expressed lncRNAs and mRNAs (Figure 3E,F). Microarray analysis indicated that 1,568 lncRNAs were differentially expressed in the sepsis group, including 695 evident up-regulation and 873 prominent down-regulation. The top 10 up-regulated and top 10 down-regulated lncRNAs are listed in Table 1. Moreover, 2,166 aberrantly expressed mRNAs also met the above criteria, in which 1,393 mRNAs were up-regulated, while 773 mRNAs were down-regulated. The top 10 most differentially expressed mRNAs (both up-regulation and down-regulation) are listed in Table 2.

Full table
Full table
Validation of microarray data by qRT-PCR
Quantitative RT-PCR indicated that the six randomly selected lncRNAs (up: Gm14832, GAS5, and Trib3; down: AK086021, Gm10497, and Ccdc104) in septic myocardium were distinctly deregulated in comparison with those in the control samples (Figure 4A,B), which were consistent with the microarray results (Figure S3). Hence, the reliability and reproducibility of the microarray analysis were further confirmed by qRT-PCR.
GO and KEGG pathway analyses
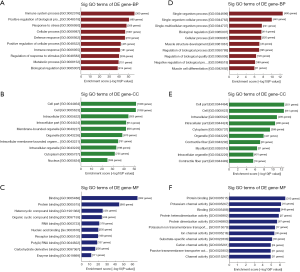
There were 2,166 distinctly deregulated mRNAs in the septic myocardium. GO analysis revealed that 1,393 up-regulated mRNAs were involved in 2,450 BP, 179 CC, 345 MF, and 773 down-regulated mRNAs who participated in 744 BP, 98 CC, and 166 MF. As shown in Figure 5A,B,C, for the upregulated mRNAs, the highest enrichment scores of the top 10 GO terms were the immune system process (GO: 0002376, 300 genes) in BP, cell part (GO: 0044464, 1,038 genes) in CC, and binding (GO: 0005488, 952 genes) in MF, respectively. However, for the downregulated mRNAs, the top enrichment in BP, CC, and MF was the single-organism process (GO: 0044699, 490 genes), cell part (GO: 0044464, 551 genes), and protein binding (GO: 0005515, 313 genes) separately (Figure 5D,E,F).
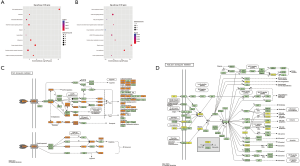
In KEGG pathway analysis, the up-regulated mRNAs were primarily associated with these pathways: TNF signaling pathway (mmu04668), herpes simplex infection (mmu05168), antigen processing and presentation (mmu04612), influenza A (mmu05164), pertussis (mmu05133), and NOD-like receptor signaling pathway (mmu04621) (Figure 6A). However, as shown in Figure 6B, breast cancer (mmu05224), axon guidance (mmu04360), central carbon metabolism in cancer (mmu05230), endocrine, and other factor-regulated calcium reabsorption (mmu04961), phosphatidylinositol-3-kinase/protein kinase B (PI3K-Akt) signaling pathway (mmu04151) and cGMP-PKG signaling pathway (mmu04022) achieved the highest enrichment scores in down-regulated mRNAs. Among the above pathways, previous studies demonstrated that the TNF signaling pathway (Figure 6C) was involved in the SMD and myocardial infarction (23,24), and the PI3K-Akt signaling pathway (Figure 6D) was related to cardiac hypertrophy, cardiac remodeling, and cardiac fibrosis (25-27).
LncRNA-mRNA co-expression network analysis
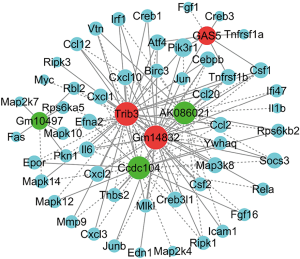
The co-expression network was created based on the above-identified six lncRNAs and their interacting mRNAs, which were extremely relevant to the TNF (40 mRNAs) and PI3K-Akt (12 mRNAs) signaling pathways. Our analysis indicated that the lncRNA-mRNA co-expression network comprised 57 network nodes and 134 connections, including 76 positive interactions and 58 negative interactions. As shown in Figure 7 and Table S2, lncRNA GM14832 interacted with 32 mRNAs; lncRNA GAS5 interacted with nine mRNAs; lncRNA Trib3 interacted with 37 mRNAs; lncRNA AK086021 interacted with 27 mRNAs; lncRNA Gm10497 interacted with seven mRNAs, and lncRNA Ccdc104 interacted with 26 mRNAs, respectively.
Discussion
In this study, microarray analysis indicated that CLP-induced sepsis led to aberrant expression of 1,568 lncRNAs (695 up-regulated and 873 down-regulated) and 2,166 mRNAs (1,393 up-regulated and 773 down-regulated) in mouse hearts, which was further validated by qRT-PCR. As far as we know, this is the first analysis of the expression profiles of lncRNAs and mRNAs by microarray technology in mouse myocardium after CLP-induced sepsis. Moreover, KEGG pathway analysis found that many aberrantly expressed mRNAs were associated with TNF and PI3K-Akt signaling pathways; and lncRNA-mRNA co-expression network analysis further demonstrated that several dysregulated lncRNAs had positive or negative interactions with numerous mRNAs that were extremely relevant to TNF and PI3K-Akt signaling pathways. Given the essential role of these two pathways during the occurrence of various heart diseases, our investigation suggested that lncRNAs and their interacting mRNAs may play a crucial role in the pathophysiological process of SMD by regulating TNF and PI3K-Akt signaling pathways.
Since the previously thought “junk genes”, lncRNAs, is now disclosed to be “functional genes” that can regulate gene expression from multiple levels, the potential role of lncRNAs in SMD has raised some concern recently. In an endotoxemia model, the overexpression of lncRNA HOTAIR in cardiomyocytes caused an enormous release of cytokine TNF-α by activating the NF-κB signaling pathway, which aggravated myocardial damage (14). Another study conducted in the sepsis model found that the overexpression of lncRNA MALAT1 in cardiomyocytes significantly increased TNF-α gene expression; however, the expression level of TNF-α was considerably reduced by blocking lncRNA MALAT1 overexpression (15). The above studies implied the involvement of lncRNAs in SMD by regulating the gene expression of TNF-α. In our research, up to 1,568 differentially expressed lncRNAs were identified in the myocardial tissue from CLP-induced septic mice, further indicating that lncRNAs indeed participate in the occurrence and development of SMD. This notion is also supported by a recent study from Chowdhury et al. (13), they found that sepsis-induced positive or negative regulation of many lncRNAs expression.
To elucidate the potential target genes that may be regulated by lncRNAs during SMD, GO and KEGG pathway analyses were used to assess the roles of all differentially expressed mRNAs. It turned out that many identified aberrantly expressed mRNAs were associated with TNF and PI3K-Akt signaling pathways. More importantly, we further prove it by lncRNA-mRNA co-expression network analysis, as several aberrantly expressed lncRNAs had positive or negative interactions with numerous mRNAs that were closely related to TNF and PI3K-Akt signaling pathways. Hence, our study indicated that lncRNAs and their interacting mRNAs might contribute to the occurrence and development of SMD and function as critical regulators of TNF and PI3K-Akt signaling pathways.
Previous studies have demonstrated that many signaling pathways were involved in the pathogenesis of SMD (14,28-31). As an essential inducer of the overwhelming inflammatory response during sepsis, TNF-α was one of the major causes of cardiac dysfunction (30,31). For instance, Yu et al. (30) found that A1 adrenoceptor activation significantly improved ventricular contractility by suppressing lipopolysaccharide (LPS)-induced TNF-α expression. Besides, LPS-stimulated inflammatory storm in macrophages was reported to be inhibited by the knockdown of lncRNA-CCL2, which alleviated organ injury due to sepsis (32). Based on these studies, and combined with our finding that differentially expressed lncRNAs and their interacting mRNAs in septic myocardium are connected to the TNF signaling pathway, it is reasonable to surmise that the TNF signaling pathway is a target for lncRNAs to modulate the pathogenesis of SMD. Another regulatory target of lncRNAs in SMD may be the PI3K-Akt signaling pathway as this pathway can regulate several intracellular signals, such as the NF-κB signaling pathway, the p53 signaling pathway, and apoptosis (29,33). Furthermore, the involvement of the PI3K-Akt signaling pathway in myocardial dysfunction has been demonstrated, including SMD, myocardial ischemia-reperfusion injury, and cardiac hypertrophy (33,34).
Autophagy, a lysosome-dependent process to degrade abnormal proteins and damaged organelles, is also reported to play a critical role in the pathogenesis of SMD (35-37). Recently, an increasing number of studies indicated that lncRNAs could modulate autophagy through multiple levels, including autophagy-related genes, phagophore nucleation, autophagosome elongation/closure, and autolysosome fusion (38,39). It has also been identified that the PI3K-Akt signaling pathway is essential in modulating autophagy and therefore preventing myocardial ischemia/reperfusion injury and myocardial fibrosis (40-42). Our microarray results did find that several autophagy-related lncRNAs, such as GAS5 and Neat1 (39,43), were abnormally expressed. Thus, it is also possible that lncRNAs play a vital role in SMD by modulating autophagy via the PI3K-Akt signaling pathway, but further studies are needed to confirm this speculation.
Although this study disclosed the expression profiles of lncRNAs and mRNAs in the myocardium derived from septic mice, TNF and PI3K-Akt signaling pathways were inferred to be the target genes regulated by aberrantly expressed lncRNAs during SMD, the limitations of our investigation should be mentioned here. First, the sample size was not large enough, which may cause a misjudgment of our results. Second, this study was only a preliminary screening in practical terms. No definite lncRNAs or mRNAs had been demonstrated to be involved in regulating TNF and PI3K-Akt signaling pathways, nor had any lncRNAs or mRNAs been identified to be associated with SMD. Hopefully, future studies would elucidate how specific lncRNAs contribute to the occurrence and development of SMD, particularly the actions of TNF and PI3K-Akt signaling pathways.
Conclusions
In summary, our study identified the profiles of aberrantly expressed lncRNAs and mRNAs in the myocardium of septic mice for the first time. According to the lncRNA-mRNA co-expression network and KEGG pathway analyses, we suggested that lncRNAs and their interacting mRNAs may be involved in the pathogenesis of SMD by regulating TNF and PI3K-Akt signaling pathways.
Acknowledgments
We would like to thank Paul McAndrew for his help in polishing the language of our paper.
Funding: This work was supported by the National Natural Science Foundation of China (81871540), the Construction Plan of Important and Weak Disciplines of Shanghai Health Commission (2016ZB0204-01), the Shanghai Science and Technology Development Funds (18RZ1429500) and the National Natural Science Foundation of China (81601682).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3830
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3830
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3830). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. SHDSYY-2018-1524) granted by the Animal Ethics Committee of Shanghai Tenth People’s Hospital, Tongji University, in compliance with the guidelines described in the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- Sun Y, Yao X, Zhang Q, et al. Beclin-1-Dependent Autophagy Protects the Heart During Sepsis. Circulation 2018;138:2247-62. [Crossref] [PubMed]
- Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med 2016;193:259-72. [Crossref] [PubMed]
- Zhou J, Tian H, Du X, et al. Population-Based Epidemiology of Sepsis in a Subdistrict of Beijing. Crit Care Med 2017;45:1168-76. [Crossref] [PubMed]
- Nasa P, Juneja D, Singh O. Severe sepsis and septic shock in the elderly: An overview. World J Crit Care Med 2012;1:23-30. [Crossref] [PubMed]
- Iwashyna TJ, Ely EW, Smith DM, et al. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA 2010;304:1787-94. [Crossref] [PubMed]
- Shankar-Hari M, Rubenfeld GD. Understanding Long-Term Outcomes Following Sepsis: Implications and Challenges. Curr Infect Dis Rep 2016;18:37. [Crossref] [PubMed]
- Hattori Y, Hattori K, Suzuki T, et al. Recent advances in the pathophysiology and molecular basis of sepsis-associated organ dysfunction: Novel therapeutic implications and challenges. Pharmacol Ther 2017;177:56-66. [Crossref] [PubMed]
- Pool R, Gomez H, Kellum JA. Mechanisms of Organ Dysfunction in Sepsis. Crit Care Clin 2018;34:63-80. [Crossref] [PubMed]
- Sun Y, Cai Y, Zang QS. Cardiac Autophagy in Sepsis. Cells 2019;8:141. [Crossref] [PubMed]
- Makara MA, Hoang KV, Ganesan LP, et al. Cardiac Electrical and Structural Changes During Bacterial Infection: An Instructive Model to Study Cardiac Dysfunction in Sepsis. J Am Heart Assoc 2016;5:e003820. [Crossref] [PubMed]
- Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv Exp Med Biol 2017;1008:47-74. [Crossref] [PubMed]
- Chowdhury IH, Narra HP, Sahni A, et al. Expression Profiling of Long Noncoding RNA Splice Variants in Human Microvascular Endothelial Cells: Lipopolysaccharide Effects In Vitro. Mediators Inflamm 2017;2017:3427461. [Crossref] [PubMed]
- Wu H, Liu J, Li W, et al. LncRNA-HOTAIR promotes TNF-alpha production in cardiomyocytes of LPS-induced sepsis mice by activating NF-kappaB pathway. Biochem Biophys Res Commun 2016;471:240-6. [Crossref] [PubMed]
- Zhuang YT, Xu DY, Wang GY, et al. IL-6 induced lncRNA MALAT1 enhances TNF-alpha expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur Rev Med Pharmacol Sci 2017;21:302-9. [PubMed]
- Zhu Y, Sun A, Meng T, et al. Protective role of long noncoding RNA CRNDE in myocardial tissues from injury caused by sepsis through the microRNA-29a/SIRT1 axis. Life Sci 2020;255:117849. [Crossref] [PubMed]
- Zhang TN, Goodwin JE, Liu B, et al. Characterization of Long Noncoding RNA and mRNA Profiles in Sepsis-Induced Myocardial Depression. Mol Ther Nucleic Acids 2019;17:852-66. [Crossref] [PubMed]
- Dai Y, Liang Z, Li Y, et al. Circulating Long Noncoding RNAs as Potential Biomarkers of Sepsis: A Preliminary Study. Genet Test Mol Biomarkers 2017;21:649-57. [Crossref] [PubMed]
- Rittirsch D, Huber-Lang MS, Flierl MA, et al. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc 2009;4:31-6. [Crossref] [PubMed]
- Chen R, Liu L, Xiao M, et al. Microarray expression profile analysis of long noncoding RNAs in premature brain injury: A novel point of view. Neuroscience 2016;319:123-33. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Royal Stat Soc Ser B 1995;57:289-300. [Crossref]
- Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279-84. [Crossref] [PubMed]
- Rahim I, Djerdjouri B, Sayed RK, et al. Melatonin administration to wild-type mice and nontreated NLRP3 mutant mice share similar inhibition of the inflammatory response during sepsis. J Pineal Res 2017. [Crossref] [PubMed]
- Zhao H, Shen R, Dong X, et al. Murine Double Minute-2 Inhibition Attenuates Cardiac Dysfunction and Fibrosis by Modulating NF-κB Pathway After Experimental Myocardial Infarction. Inflammation 2017;40:232-9. [Crossref] [PubMed]
- Weeks KL, Bernardo BC, Ooi JYY, et al. The IGF1-PI3K-Akt Signaling Pathway in Mediating Exercise-Induced Cardiac Hypertrophy and Protection. Adv Exp Med Biol 2017;1000:187-210. [Crossref] [PubMed]
- Yang W, Wu Z, Yang K, et al. BMI1 promotes cardiac fibrosis in ischemia-induced heart failure via the PTEN-PI3K/Akt-mTOR signaling pathway. Am J Physiol Heart Circ Physiol 2019;316:H61-H69. [Crossref] [PubMed]
- Wang HB, Hang S, Xu M, et al. Galangin ameliorates cardiac remodeling via the MEK1/2-ERK1/2 and PI3K-AKT pathways. J Cell Physiol 2019;234:15654-67. [Crossref] [PubMed]
- Piquereau J, Godin R, Deschênes S, et al. Protective role of PARK2/Parkin in sepsis-induced cardiac contractile and mitochondrial dysfunction. Autophagy 2013;9:1837-51. [Crossref] [PubMed]
- Fan TT, Feng XY, Yang YZ, et al. Downregulation of PI3K-gamma in a mouse model of sepsis-induced myocardial dysfunction. Cytokine 2017;96:208-16. [Crossref] [PubMed]
- Yu X, Jia B, Wang F, et al. alpha(1) adrenoceptor activation by norepinephrine inhibits LPS-induced cardiomyocyte TNF-alpha production via modulating ERK1/2 and NF-kappaB pathway. J Cell Mol Med 2014;18:263-73. [Crossref] [PubMed]
- Zhai J, Guo Y. Paeoniflorin attenuates cardiac dysfunction in endotoxemic mice via the inhibition of nuclear factor-kappaB. Biomed Pharmacother 2016;80:200-6. [Crossref] [PubMed]
- Jia Y, Li Z, Cai W, et al. SIRT1 regulates inflammation response of macrophages in sepsis mediated by long noncoding RNA. Biochim Biophys Acta Mol Basis Dis 2018;1864:784-92. [Crossref] [PubMed]
- An R, Zhao L, Xi C, et al. Melatonin attenuates sepsis-induced cardiac dysfunction via a PI3K/Akt-dependent mechanism. Basic Res Cardiol 2016;111:8. [Crossref] [PubMed]
- Hernández-Reséndiz S, Palma-Flores C, De Los Santos S, et al. Reduction of no-reflow and reperfusion injury with the synthetic 17beta-aminoestrogen compound Prolame is associated with PI3K/Akt/eNOS signaling cascade. Basic Res Cardiol 2015;110:1. [Crossref] [PubMed]
- Hsiao HW, Tsai KL, Wang LF, et al. The decline of autophagy contributes to proximal tubular dysfunction during sepsis. Shock 2012;37:289-96. [Crossref] [PubMed]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010;12:823-30. [Crossref] [PubMed]
- Watanabe E, Muenzer JT, Hawkins WG, et al. Sepsis induces extensive autophagic vacuolization in hepatocytes: a clinical and laboratory-based study. Lab Invest 2009;89:549-61. [Crossref] [PubMed]
- Yang L, Wang H, Shen Q, et al. Long noncoding RNAs involved in autophagy regulation. Cell Death Dis 2017;8:e3073. [Crossref] [PubMed]
- Zhang J, Wang P, Wan L, et al. The emergence of noncoding RNAs as Heracles in autophagy. Autophagy 2017;13:1004-24. [Crossref] [PubMed]
- Xiao T, Luo J, Wu Z, et al. Effects of hydrogen sulfide on myocardial fibrosis and PI3K/AKT1-regulated autophagy in diabetic rats. Mol Med Rep 2016;13:1765-73. [Crossref] [PubMed]
- Xuan F, Jian J, Lin X, et al. 17-Methoxyl-7-Hydroxy-Benzene-Furanchalcone Ameliorates Myocardial Ischemia/Reperfusion Injury in Rat by Inhibiting Apoptosis and Autophagy Via the PI3K-Akt Signal Pathway. Cardiovasc Toxicol 2017;17:79-87. [Crossref] [PubMed]
- Zheng Y, Gu S, Li X, et al. Berbamine postconditioning protects the heart from ischemia/reperfusion injury through modulation of autophagy. Cell Death Dis 2017;8:e2577. [Crossref] [PubMed]
- Ma M, Hui J, Zhang Q-y, et al. Long noncoding RNA nuclear-enriched abundant transcript 1 inhibition blunts myocardial ischemia reperfusion injury via autophagic flux arrest and apoptosis in streptozotocin-induced diabetic rats. Atherosclerosis 2018;277:113-22. [Crossref] [PubMed]


