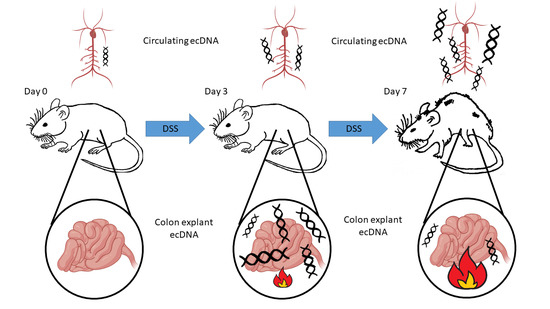
Extracellular DNA Correlates with Intestinal Inflammation in Chemically Induced Colitis in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Induction of Colitis
Administration of PAD4 Inhibitors
2.3. Assessment of Colitis Severity
2.4. Isolation of Samples
2.5. Protein Concentration Measurement
2.6. Colonic Explant Preparation
2.7. Animal Endoscopy
2.8. ecDNA Isolation and Measurement
2.9. Real-Time PCR
2.10. MPO Activity Measurement
2.11. TNF-α Measurement
2.12. Determination of DNase Activity
2.13. Flow Cytometry
2.14. Massively Parallel Sequencing of Plasma ecDNA
2.15. Bioinformatics Processing
2.16. Whole-Mount Immunostaining of the Colon
2.17. Statistical Analysis
3. Results
3.1. Dextran Sulfate Sodium (DSS) Induced Intestinal Inflammation
3.2. Total ecDNA Is Increased in Plasma but Not in Colon Explant Medium
3.3. Specific Fragmentation and Origin of Plasma ecDNA Related to Colitis Severity
3.4. PAD4 Is Increased in Colonic Tissue of Mice with Severe Colitis
3.5. Inhibition of NETosis Partially Ameliorated Colitis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2018, 390, 2769–2778. [Google Scholar] [CrossRef]
- Pieczyńska, J.; Prescha, A.; Zabłocka-Słowińska, K.; Neubauer, K.; Smereka, A.; Grajeta, H.; Biernat, J.; Paradowski, L. Occurrence of dietary risk factors in inflammatory bowel disease: Influence on the nutritional status of patients in clinical remission. Adv. Clin. Exp. Med. 2019, 28, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.; Totino, V.; Cacciotti, F.; Iebba, V.; Neroni, B.; Bonfiglio, G.; Trancassini, M.; Passariello, C.; Pantanella, F.; Schippa, S. Rebuilding the gut microbiota ecosystem. Int. J. Environ. Res. Public Health 2018, 15, 1679. [Google Scholar]
- De Meij, T.G.J.; De Groot, E.F.J.; Peeters, C.F.W.; De Boer, N.K.H.; Kneepkens, C.M.F.; Eck, A.; Benninga, M.A.; Savelkoul, P.H.M.; Van Bodegraven, A.A.; Budding, A.E. Variability of core microbiota in newly diagnosed treatment-naïve paediatric inflammatory bowel disease patients. PLoS ONE 2018, 13, e0197649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.; Yang, J.; Ye, W.; Zhang, Y.; Tang, C.; Li, X.; Zhou, X.; Xie, Y. Urban–rural environmental exposure during childhood and subsequent risk of inflammatory bowel disease: A meta-analysis. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 591–602. [Google Scholar] [CrossRef] [PubMed]
- O’Driscoll, L. Extracellular nucleic acids and their potential as diagnostic, prognostic and predictive biomarkers. Anticancer Res. 2007, 27, 1257–1265. [Google Scholar]
- Zhong, X.Y.; Burk, M.R.; Troeger, C.; Kang, A.; Holzgreve, W.; Hahn, S. Fluctuation of maternal and fetal free extracellular circulatory DNA in maternal plasma. Obstet. Gynecol. 2000, 96, 991–996. [Google Scholar]
- Lui, Y.Y.N.; Woo, K.-S.; Wang, A.Y.M.; Yeung, C.-K.; Li, P.K.T.; Chau, E.; Ruygrok, P.; Lo, Y.M.D. Origin of plasma cell-free DNA after solid organ transplantation. Clin. Chem. 2003, 49, 495–496. [Google Scholar] [CrossRef] [Green Version]
- Nishimoto, S.; Fukuda, D.; Sata, M. Emerging roles of Toll-like receptor 9 in cardiometabolic disorders. Inflamm. Regen. 2020, 40, 1–13. [Google Scholar] [CrossRef]
- Krieg, A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002, 20, 709–760. [Google Scholar] [CrossRef]
- Sharma, S.; Fitzgerald, K.A. Innate immune sensing of DNA. PLoS Pathog. 2011, 7, e1001310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avriel, A.; Wiessman, M.P.; Almog, Y.; Perl, Y.; Novack, V.; Galante, O.; Klein, M.; Pencina, M.J.; Douvdevani, A. Admission cell free DNA levels predict 28-day mortality in patients with severe sepsis in intensive care. PLoS ONE 2014, 9, e100514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clementi, A.; Virzi, G.M.; Brocca, A.; Pastori, S.; de Cal, M.; Marcante, S.; Granata, A.; Ronco, C. The Role of Cell-Free Plasma DNA in Critically Ill Patients with Sepsis. Blood Purif. 2016, 41, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Schneck, E.; Samara, O.; Koch, C.; Hecker, A.; Padberg, W.; Lichtenstern, C.; Weigand, M.A.; Uhle, F. Plasma DNA and RNA differentially impact coagulation during abdominal sepsis-an explorative study. J. Surg. Res. 2017, 210, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.I.; Soliman, R.A.; Samir, S. Cell Free DNA and Procalcitonin as Early Markers of Complications in ICU Patients with Multiple Trauma and Major Surgery. Clin. Lab. 2016, 62, 2395–2404. [Google Scholar] [CrossRef]
- Nishimoto, S.; Fukuda, D.; Higashikuni, Y.; Tanaka, K.; Hirata, Y.; Murata, C.; Kim-Kaneyama, J.R.; Sato, F.; Bando, M.; Yagi, S.; et al. Obesity-induced DNA released from adipocytes stimulates chronic adipose tissue inflammation and insulin resistance. Sci. Adv. 2016, 2, e1501332. [Google Scholar] [CrossRef] [Green Version]
- Koike, Y.; Uchida, K.; Tanaka, K.; Ide, S.; Otake, K.; Okita, Y.; Inoue, M.; Araki, T.; Mizoguchi, A.; Kusunoki, M. Dynamic pathology for circulating free DNA in a dextran sodium sulfate colitis mouse model. Pediatr. Surg. Int. 2014, 30, 1199–1206. [Google Scholar] [CrossRef]
- Maronek, M.; Gromova, B.; Liptak, R.; Klimova, D.; Cechova, B.; Gardlik, R. Extracellular DNA is Increased in Dextran Sulphate Sodium-Induced Colitis in Mice. Folia Biol. (Praha) 2018, 64, 167–172. [Google Scholar]
- Sipos, F.; Műzes, G.; Fűri, I.; Spisák, S.; Wichmann, B.; Germann, T.M.; Constantinovits, M.; Krenács, T.; Tulassay, Z.; Molnár, B. Intravenous administration of a single-dose free-circulating DNA of colitic origin improves severe murine DSS-colitis. Pathol. Oncol. Res. 2014, 20, 867–877. [Google Scholar]
- Műzes, G.; Kiss, A.L.; Tulassay, Z.; Sipos, F. Cell-free DNA-induced alteration of autophagy response and TLR9-signaling: Their relation to amelioration of DSS-colitis. Comp. Immunol. Microbiol. Infect. Dis. 2017, 52, 48–57. [Google Scholar] [CrossRef]
- Constantinovits, M.; Sipos, F.; Kiss, A.L.; Műzes, G. Preconditioning with cell-free DNA prevents DSS-colitis by promoting cell protective autophagy. J. Investig. Med. 2020, 68, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Vaibhav, K.; Braun, M.; Alverson, K.; Khodadadi, H.; Kutiyanawalla, A.; Ward, A.; Banerjee, C.; Sparks, T.; Malik, A.; Rashid, M.H.; et al. Neutrophil extracellular traps exacerbate neurological deficits after traumatic brain injury. Sci. Adv. 2020, 6, eaax8847. [Google Scholar] [CrossRef] [PubMed]
- Meijenfeldt, F.A.; von Jenne, C.N. Netting Liver Disease: Neutrophil Extracellular Traps in the Initiation and Exacerbation of Liver Pathology. Semin. Thromb. Hemost. 2020, 46, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Rivera, C.; Carlucci, P.M.; Goel, R.R.; James, E.; Brooks, S.R.; Rims, C.; Hoffmann, V.; Fox, D.A.; Buckner, J.H.; Kaplan, M.J. Neutrophil extracellular traps mediate articular cartilage damage and enhance cartilage component immunogenicity in rheumatoid arthritis. JCI Insight 2020, 5, e139388. [Google Scholar]
- Lin, E.Y.H.; Lai, H.J.; Cheng, Y.K.; Leong, K.Q.; Cheng, L.C.; Chou, Y.C.; Peng, Y.C.; Hsu, Y.H.; Chiang, H. Sen Neutrophil extracellular traps impair intestinal barrier function during experimental colitis. Biomedicines 2020, 8, 275. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, H.; Onuma, A.E.; Tsung, A. Neutrophil Elastase and Neutrophil Extracellular Traps in the Tumor Microenvironment. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2020; Volume 1263, pp. 13–23. [Google Scholar]
- Yang, L.; Liu, L.; Zhang, R.; Hong, J.; Wang, Y.; Wang, J.; Zuo, J.; Zhang, J.; Chen, J.; Hao, H. IL-8 mediates a positive loop connecting increased neutrophil extracellular traps (NETs) and colorectal cancer liver metastasis. J. Cancer 2020, 11, 4384–4396. [Google Scholar] [CrossRef]
- Laukova, L.; Konecna, B.; Babickova, J.; Wagnerova, A.; Meliskova, V.; Vlkova, B.; Celec, P. Exogenous deoxyribonuclease has a protective effect in a mouse model of sepsis. Biomed. Pharmacother. 2017, 93, 8–16. [Google Scholar]
- Vokálová, L.; Lauková, L.; Čonka, J.; Melišková, V.; Borbélyová, V.; Bábíčková, J.; Tóthová, L.; Hodosy, J.; Vlková, B.; Celec, P. Deoxyribonuclease partially ameliorates thioacetamide-induced hepatorenal injury. Am. J. Physiol. Liver Physiol. 2017, 312, G457–G463. [Google Scholar] [CrossRef]
- Mondal, S.; Thompson, P.R. Protein Arginine Deiminases (PADs): Biochemistry and Chemical Biology of Protein Citrullination. Acc. Chem. Res. 2019, 52, 818–832. [Google Scholar]
- Mutua, V.; Gershwin, L.J. A Review of Neutrophil Extracellular Traps (NETs) in Disease: Potential Anti-NETs Therapeutics. Clin. Rev. Allergy Immunol. 2020, 1, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.N.; Czajka, A.; Cunningham, P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion 2016, 29, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Mei, Y.; Dong, W.; Wang, J.; Huang, F.; Wu, J. Evaluation of protein arginine deiminase-4 inhibitor in TNBS- induced colitis in mice. Int. Immunopharmacol. 2020, 84, 106583. [Google Scholar] [PubMed]
- Kim, J.J.; Shajib, M.S.; Manocha, M.M.; Khan, W.I. Investigating intestinal inflammation in DSS-induced model of IBD. J. Vis. Exp. 2012, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinallo, V.; Marafini, I.; Di Fusco, D.; Laudisi, F.; Franzè, E.; Di Grazia, A.; Figliuzzi, M.M.; Caprioli, F.; Stolfi, C.; Monteleone, I.; et al. Neutrophil Extracellular Traps Sustain Inflammatory Signals in Ulcerative Colitis. J. Crohns. Colitis 2019, 13, 772–784. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Malíčková, K.; Duricová, D.; Bortlík, M.; Hrušková, Z.; Svobodová, B.; Machková, N.; Komárek, V.; Fučíková, T.; Janatková, I.; Zima, T.; et al. Impaired deoxyribonuclease I activity in patients with inflammatory bowel diseases. Autoimmune Dis. 2011, 2011, 945861. [Google Scholar] [CrossRef] [Green Version]
- Babraham, A. Bioinformatics. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 31 October 2020).
- Yuzhalin, A.E. Citrullination in cancer. Cancer Res. 2019, 79, 1274–1284. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Luo, X.; Qian, J.; Pang, X.; Song, J.; Qian, G.; Chen, J.; Chen, S. FastUniq: A Fast De Novo Duplicates Removal Tool for Paired Short Reads. PLoS ONE 2012, 7, e52249. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wang, D.; Yao, H.; Doret, P.; Hao, G.; Shen, Q.; Qiu, H.; Zhang, X.; Wang, Y.; Chen, G.; et al. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene 2010, 29, 3153–3162. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [PubMed] [Green Version]
- Babickova, J.; Conka, J.; Janovicova, L.; Boris, M.; Konecna, B.; Gardlik, R. Extracellular DNA as a Prognostic and Therapeutic Target in Mouse Colitis under DNase I Treatment. Folia Biol. (Praha) 2018, 64, 10–15. [Google Scholar] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubiritova, Z.; Radvanszky, J.; Gardlik, R. Cell-Free Nucleic Acids and their Emerging Role in the Pathogenesis and Clinical Management of Inflammatory Bowel Disease. Int. J. Mol. Sci. 2019, 20, 3662. [Google Scholar]
- Chumanevich, A.A.; Causey, C.P.; Knuckley, B.A.; Jones, J.E.; Poudyal, D.; Chumanevich, A.P.; Davis, T.; Matesic, L.E.; Thompson, P.R.; Hofseth, L.J. Suppression of colitis in mice by Cl-amidine: A novel peptidylarginine deiminase inhibitor. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G929–G938. [Google Scholar] [CrossRef] [PubMed]
- Dreyton, C.; Jones, J.; Knuckley, B.; Subramanian, V.; Anderson, E.; Brown, S.; Fernandez-Vega, V.; Eberhart, C.; Spicer, T.; Zuhl, A.; et al. Optimization and characterization of a pan protein arginine deiminase (PAD) inhibitor. In Probe Reports from the NIH Molecular Libraries Program [Internet]; National Center for Biotechnology Information: Bethesda, MD, USA, 2013. [Google Scholar]









| Category | Score | Description |
|---|---|---|
| Translucency | 0 | Vascularization fully visible |
| 1 | Vascularization partially visible | |
| 2 | Vascularization not visible | |
| Fibrin | 0 | No fibrin is present in the mucosa |
| 1 | Small fibrin deposits in the mucosa | |
| 2 | Large fibrin deposits in the mucosa | |
| Bleeding | 0 | No bleeding |
| 1 | Several sites of mucosal bleeding | |
| 2 | Many sites of mucosal bleeding, may obstruct camera of the endoscope, bleeding may start spontaneously or as a reaction to contact with endoscope, blood may directly flow out of the anus | |
| 3 | Profound mucosal bleeding, usually obstructs camera of the endoscope, bleeding often starts spontaneously and blood usually flows out of the anus | |
| Reddening | 0 | No reddening visible |
| 1 | Several sites of mucosal reddening | |
| 2 | Many sites of mucosal reddening |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maronek, M.; Gromova, B.; Liptak, R.; Konecna, B.; Pastorek, M.; Cechova, B.; Harsanyova, M.; Budis, J.; Smolak, D.; Radvanszky, J.; et al. Extracellular DNA Correlates with Intestinal Inflammation in Chemically Induced Colitis in Mice. Cells 2021, 10, 81. https://doi.org/10.3390/cells10010081
Maronek M, Gromova B, Liptak R, Konecna B, Pastorek M, Cechova B, Harsanyova M, Budis J, Smolak D, Radvanszky J, et al. Extracellular DNA Correlates with Intestinal Inflammation in Chemically Induced Colitis in Mice. Cells. 2021; 10(1):81. https://doi.org/10.3390/cells10010081
Chicago/Turabian StyleMaronek, Martin, Barbora Gromova, Robert Liptak, Barbora Konecna, Michal Pastorek, Barbora Cechova, Maria Harsanyova, Jaroslav Budis, David Smolak, Jan Radvanszky, and et al. 2021. "Extracellular DNA Correlates with Intestinal Inflammation in Chemically Induced Colitis in Mice" Cells 10, no. 1: 81. https://doi.org/10.3390/cells10010081