Immune-Response Patterns and Next Generation Sequencing Diagnostics for the Detection of Mycoses in Patients with Septic Shock—Results of a Combined Clinical and Experimental Investigation
Abstract
:1. Introduction
2. Results
2.1. Patient’s Characteristics
2.2. Fungal Pathogens and Infection Sites
Culture-Based Microbiological Diagnostics
2.3. NGS-Based Microbiological Diagnostics
2.3.1. Confirmation of Culture-Based Diagnostics of Candidemia by Fungal SIQ Score
2.3.2. NGS-Based Diagnosis of Candidemia in Patients with Suggested Infection Based on Microbiological Identification of Candida spp. in Other Than Blood Culture
2.3.3. Detection of Invasive Mycoses by Fungal SIQ Score Even in Those Patients Which Have So Far Been Classified as Colonized Based on Culture-Based Diagnostic Procedures
2.4. Antifungal Therapy
2.5. (1,3)-β-d-Glucan (BG)
2.6. Galactomannan (GM)
2.7. Anti-Candida Antibody Titer
2.8. Inflammation as Well as Infection Marker Levels
2.9. Conserved Host-Response Patterns during Epithelial Infections with Candida spp.
3. Discussion
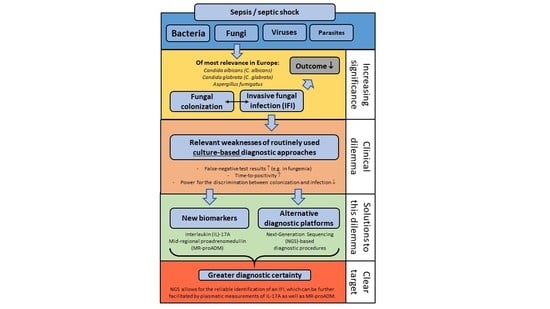
3.1. Invasive Fungal Infections—A Clinical Dilemma
3.2. Solutions to the Fungal Dilemma
3.3. Next-Generation Sequencing (NGS) Diagnostics of Fungal Infections
3.4. Fungal Biomarkers for the Diagnosis of Fungal Infections
3.5. Immune Monitoring as a Diagnostic Tool in Fungal Infections
3.6. Limitations
4. Materials and Methods
4.1. Study Design
4.2. Immunoassays
4.3. Flowcytometry
4.4. Clinical Microbiology
4.4.1. Blood Culture
4.4.2. Culture-Based Diagnostic Procedures in Tracheal Secretion, Wound Swabs, and Drainage Fluids
4.4.3. Group Definitions
4.4.4. Anti-Candida-Antibody Titer
4.5. Next Generation Sequencing (NGS)-Based Diagnostic Procedures
4.5.1. Plasma Preparation and Nucleic Acid Isolation
4.5.2. Preparation of NGS Libraries and Sequencing
4.5.3. Bioinformatics
4.6. Gene Expression Analysis of Infected Reconstituted Human Epithelia (RHE)
4.6.1. Cell Culture Model
4.6.2. RNA-Seq
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AATI | HS NGS Fragment Analysis Kit and the Fragment Analyzer instrument |
| ADM | Adrenomedullin |
| APACHE II | Acute Physiology Health Evaluation score |
| ARDS | Acute respiratory distress syndrome |
| AST | automated antibiotic susceptibility testing |
| BALF | Bronchoalveolar lavage fluid |
| BC | Blood culture |
| BMI | Body mass index |
| BG | β-d-glucan |
| bp | Base pair |
| C. albicans | Candida albicans |
| C. glabrata | Candida glabrata |
| Candida spp. | Candida species |
| cfDNA | Cell-free DNA |
| CFG | Caspofungin |
| CLR | C-type leptin receptors |
| COPD | Chronic obstructive pulmonary disease |
| CRP | C-reactive protein |
| CVC | Central venous catheter |
| DC | Dendritic cells |
| DNA | Deoxyribonucleic acid |
| DOR | Doripenem |
| ELISA | Enzyme-linked Immunosorbent Assay |
| EORTC | European Organisation for Research and Treatment of Cancer |
| FLC | Fluconazole |
| GIT | Gastro intestinal tract |
| GM | Galactomannan |
| IA | Invasive Aspergillosis |
| ICU | Intensive Care Unit |
| IPM:CIL | Imipenem/Cilastatin |
| INF-γ | Interferon gamma |
| IL | Interleukin |
| IL-17A | Interleukin-17A |
| MEM | Meropenem |
| MR-proADM | Mid-regional proadrenomedullin |
| MSG | National Institute of Allergy and Infectious Disease Mycoses Study Group |
| na | Not available |
| nd | Not detectable |
| NGS | Next-generation sequencing |
| NYHA | New York Heart association score |
| PAMP | aminotemrinal peptide of proadrenomedullin |
| PAMPs | Pathogen associated molecular patterns |
| PCR | Polymerase chain reaction |
| PCT | Procalcitonin |
| PRR | Pattern recognition receptors |
| RHE | Reconstituted human epithelia |
| RPKM | Reads per kilobase of exon model per million |
| SAPS II | Simplified Acute Physiology Score |
| SIQ | Sepsis indicating quantifier |
| SOFA | Sequential Organ Failure Assessment Score |
| spp. | Species |
| Treg | Regulatory T-cell |
| TBC | Inhaled Tobramycine |
| TLR | Toll-like receptor |
| TRACE | Time Resolved Amplified Cryptate Emission |
| TS | Tracheal secretion |
| TZP | Piperacilline/Tazobactam |
| VAC | Vancomycin |
| VAS | Vascular artery surgery |
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.; Sibbald, W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A. Management of sepsis. N. Engl. J. Med. 2006, 355, 1699–1713. [Google Scholar] [CrossRef] [PubMed]
- Waterer, G.W.; ElBahlawan, L.; Quasney, M.W.; Zhang, Q.; Kessler, L.A.; Wunderink, R.G. Heat shock protein 70-2+ 1267 AA homozygotes have an increased risk of septic shock in adults with community-acquired pneumonia. Crit. Care Med. 2003, 31, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Eggimann, P.; Garbino, J.; Pittet, D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 2003, 3, 685–702. [Google Scholar] [CrossRef]
- Jojima, H. Early diagnosis and treatment of pulmonary opportunistic infection by using polymerase chain reaction and β-glucan in patients with hematological neoplasms. Kurume Med. J. 2001, 48, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Aguado, I.; Calvo, C.; Wilhelmi, I.; Pablo-Hernando, M.E.; Medina, M.J.; Saez-Nieto, J.A.; Cabrerizo, M. Sepsis and meningitis caused by pasteurella multocida and echovirus 9 in a neonate. Pediatr. Infect. Dis. J. 2014, 33, 1308–1309. [Google Scholar] [CrossRef] [PubMed]
- Kerwat, K.; Rolfes, C.; Wulf, H. Fungal infections in the intensive care unit. AINS 2011, 46, 744–745. [Google Scholar] [PubMed]
- Kett, D.H.; Azoulay, E.; Echeverria, P.M.; Vincent, J.L. Candida bloodstream infections in intensive care units: Analysis of the extended prevalence of infection in intensive care unit study. Crit. Care Med. 2011, 39, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Solomkin, J.S.; Mazuski, J.E.; Bradley, J.S.; Rodvold, K.A.; Goldstein, E.J.; Baron, E.J.; O’Neill, P.J.; Chow, A.W.; Dellinger, E.P.; Eachempati, S.R.; et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: Guidelines by the surgical infection society and the infectious diseases society of america. Surg. Infect. (Larchmt) 2010, 11, 79–109. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Marchetti, M.; Chakrabarti, A.; Colizza, S.; Garnacho-Montero, J.; Kett, D.H.; Munoz, P.; Cristini, F.; Andoniadou, A.; Viale, P.; et al. A research agenda on the management of intra-abdominal candidiasis: Results from a consensus of multinational experts. Intensive Care Med. 2013, 39, 2092–2106. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E.; Costa, A.; Fasce, R.; Molinari, M.P.; Rosso, R.; Pallavicini, F.B.; Viscoli, C. Epidemiological trends in nosocomial candidemia in intensive care. BMC Infect. Dis. 2006, 10, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.S.; Mannino, D.M.; Eaton, S.; Moss, M. The epidemiology of sepsis in the united states from 1979 through 2000. N. Engl. J. Med. 2003, 348, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstern, C.; Herold, C.; Mieth, M.; Brenner, T.; Decker, S.; Busch, C.J.; Hofer, S.; Zimmermann, S.; Weigand, M.A.; Bernhard, M. Relevance of Candida and other mycoses for morbidity and mortality in severe sepsis and septic shock due to peritonitis. Mycoses 2015, 58, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; Sakr, Y.; Sprung, C.L.; Ranieri, V.M.; Reinhart, K.; Gerlach, H.; Moreno, R.; Carlet, J.; Le Gall, J.R.; Payen, D.; et al. Sepsis in european intensive care units: Results of the soap study. Crit. Care Med. 2006, 34, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Shorr, A.F.; Gupta, V.; Sun, X.; Johannes, R.S.; Spalding, J.; Tabak, Y.P. Burden of early-onset candidemia: Analysis of culture-positive bloodstream infections from a large U.S. Database. Crit. Care Med. 2009, 37, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Trof, R.J.; Beishuizen, A.; Debets-Ossenkopp, Y.J.; Girbes, A.R.; Groeneveld, A.B. Management of invasive pulmonary aspergillosis in non-neutropenic critically ill patients. Intensive Care Med. 2007, 33, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Zaoutis, T.E.; Argon, J.; Chu, J.; Berlin, J.A.; Walsh, T.J.; Feudtner, C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the united states: A propensity analysis. Clin. Infect. Dis. 2005, 41, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Combes, A.; Mokhtari, M.; Couvelard, A.; Trouillet, J.L.; Baudot, J.; Henin, D.; Gibert, C.; Chastre, J. Clinical and autopsy diagnoses in the intensive care unit: A prospective study. Arch. Intern. Med. 2004, 164, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Mort, T.C.; Yeston, N.S. The relationship of pre mortem diagnoses and post mortem findings in a surgical intensive care unit. Crit. Care Med. 1999, 27, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Silfvast, T.; Takkunen, O.; Kolho, E.; Andersson, L.C.; Rosenberg, P. Characteristics of discrepancies between clinical and autopsy diagnoses in the intensive care unit: A 5-year review. Intensive Care Med. 2003, 29, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Kimura, M.; Araoka, H.; Taniguchi, S.; Yoneyama, A. Is initial serum (1,3)-β-d-glucan truly associated with mortality in patients with candidaemia? Clin. Microbiol. Infect. 2016, 22, 576. [Google Scholar] [CrossRef] [PubMed]
- Garey, K.W.; Rege, M.; Pai, M.P.; Mingo, D.E.; Suda, K.J.; Turpin, R.S.; Bearden, D.T. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: A multi-institutional study. Clin. Infect. Dis. 2006, 43, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E.; Ansaldi, F.; Merelli, M.; Trucchi, C.; de Pascale, G.; Diaz-Martin, A.; Luzzati, R.; Rosin, C.; Lagunes, L.; et al. A multicenter study of septic shock due to candidemia: Outcomes and predictors of mortality. Intensive Care Med. 2014, 40, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Avni, T.; Leibovici, L.; Paul, M. Pcr diagnosis of invasive candidiasis: Systematic review and meta-analysis. J. Clin. Microbiol. 2011, 49, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Garnacho-Montero, J.; Calandra, T.; Kullberg, B.; Dimopoulos, G.; Azoulay, E.; Chakrabarti, A.; Kett, D.; Leon, C.; Ostrosky-Zeichner, L.; et al. Intensive care medicine research agenda on invasive fungal infection in critically ill patients. Intensive Care Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Boonsarngsuk, V.; Niyompattama, A.; Teosirimongkol, C.; Sriwanichrak, K. False-positive serum and bronchoalveolar lavage aspergillus galactomannan assays caused by different antibiotics. Scand. J. Infect. Dis. 2010, 42, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Metan, G. The interaction between piperacillin-tazobactam and aspergillus galactomannan antigenemia assay: Is the story over? Infection 2013, 41, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Naglik, J.R. Mucosal immunity and Candida albicans infection. Clin. Dev. Immunol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Brunkhorst, F.M.; Oppert, M.; Marx, G.; Bloos, F.; Ludewig, K.; Putensen, C.; Nierhaus, A.; Jaschinski, U.; Meier-Hellmann, A.; Weyland, A.; et al. Effect of empirical treatment with moxifloxacin and meropenem vs meropenem on sepsis-related organ dysfunction in patients with severe sepsis: A randomized trial. JAMA 2012, 307, 2390–2399. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.; Brunkhorst, F.M.; Bone, H.G.; Brunkhorst, R.; Gerlach, H.; Grond, S.; Gruendling, M.; Huhle, G.; Jaschinski, U.; John, S.; et al. Epidemiology of sepsis in germany: Results from a national prospective multicenter study. Intensive Care Med. 2007, 33, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.P.; Keller, P.M.; Baier, M.; Hagel, S.; Pletz, M.W.; Brunkhorst, F.M. Quality of blood culture testing—A survey in intensive care units and microbiological laboratories across four european countries. Crit. Care 2013, 17. [Google Scholar] [CrossRef] [PubMed]
- Kirn, T.J.; Weinstein, M.P. Update on blood cultures: How to obtain, process, report, and interpret. Clin. Microbiol. Infect. 2013, 19, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Bassetti, M.; Calandra, T.; Garbino, J.; Kullberg, B.J.; Lortholary, O.; Meersseman, W.; Akova, M.; Arendrup, M.C.; Arikan-Akdagli, S.; et al. Escmid guideline for the diagnosis and management of Candida diseases 2012: Non-neutropenic adult patients. Clin. Microbiol. Infect. 2012, 18, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Leon, C.; Ruiz-Santana, S.; Saavedra, P.; Almirante, B.; Nolla-Salas, J.; Alvarez-Lerma, F.; Garnacho-Montero, J.; Leon, M.A. A bedside scoring system “Candida score” for early antifungal treatment in nonneutropenic critically ill patients with Candida colonization. Crit. Care Med. 2006, 34, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G.; Lambiotte, F.; Thevenin, D.; Lemaire, C.; Parmentier, E.; Devos, P.; Leroy, O. Evaluation of “Candida score” in critically ill patients: A prospective, multicenter, observational, cohort study. Ann. Intensive Care 2011, 1, 50. [Google Scholar] [CrossRef] [PubMed]
- Borg-von Zepelin, M.; Kunz, L.; Ruchel, R.; Reichard, U.; Weig, M.; Gross, U. Epidemiology and antifungal susceptibilities of Candida spp. To six antifungal agents: Results from a surveillance study on fungaemia in germany from july 2004 to august 2005. J. Antimicrob. Chemother. 2007, 60, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.; Guidry, C.; Politano, A.; Rosenberger, L.; McLeod, M.; Hranjec, T.; Sawyer, R. Aspergillus infections in transplant and non-transplant surgical patients. Surg. Infect. (Larchmt) 2014, 15, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Anaissie, E.J.; Denning, D.W.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Segal, B.H.; Steinbach, W.J.; Stevens, D.A.; et al. Treatment of aspergillosis: Clinical practice guidelines of the infectious diseases society of america. Clin. Infect. Dis. 2008, 46, 327–360. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.H.; Fang, X.M.; Fang, Q.; Wu, X.M.; Jin, Y.H.; Wang, J.L.; Guo, Q.L.; Gu, M.N.; Xu, Q.P.; Wang, D.X.; et al. Impact of invasive fungal infection on outcomes of severe sepsis: A multicenter matched cohort study in critically ill surgical patients. Crit. Care 2008, 12, R5. [Google Scholar] [CrossRef] [PubMed]
- Lewejohann, J.; Hansen, M.; Zimmermann, C.; Muhl, E.; Bruch, H.P. Recurrent Candida sepsis with prolonged respiratory failure and severe liver dysfunction. Mycoses 2005, 48, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Markgraf, R.; Deutschinoff, G.; Pientka, L.; Scholten, T.; Lorenz, C. Performance of the score systems acute physiology and chronic health evaluation II and III at an interdisciplinary intensive care unit, after customization. Crit. Care 2001, 5, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Dupont, H.; Bourichon, A.; Paugam-Burtz, C.; Mantz, J.; Desmonts, J.M. Can yeast isolation in peritoneal fluid be predicted in intensive care unit patients with peritonitis? Crit. Care Med. 2003, 31, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Montravers, P.; Mira, J.P.; Gangneux, J.P.; Leroy, O.; Lortholary, O.; AmarCand study group. A multicentre study of antifungal strategies and outcome of Candida spp. Peritonitis in intensive-care units. Clin. Microbiol. Infect. 2011, 17, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.; Catalan, M.; del Palacio-Perez-Medel, A.; Lora, D.; Montejo, J.C.; Cuetara, M.S.; Moragues, M.D.; Ponton, J.; del Palacio, A. A prospective comparison of galactomannan in bronchoalveolar lavage fluid for the diagnosis of pulmonary invasive aspergillosis in medical patients under intensive care: Comparison with the diagnostic performance of galactomannan and of (1→3)-β-d-glucan chromogenic assay in serum samples. Clin. Microbiol. Infect. 2011, 17, 1053–1060. [Google Scholar] [PubMed]
- Fukuda, T.; Boeckh, M.; Carter, R.A.; Sandmaier, B.M.; Maris, M.B.; Maloney, D.G.; Martin, P.J.; Storb, R.F.; Marr, K.A. Risks and outcomes of invasive fungal infections in recipients of allogeneic hematopoietic stem cell transplants after nonmyeloablative conditioning. Blood 2003, 102, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Montravers, P.; Dupont, H.; Gauzit, R.; Veber, B.; Auboyer, C.; Blin, P.; Hennequin, C.; Martin, C. Candida as a risk factor for mortality in peritonitis. Crit. Care Med. 2006, 34, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Fiore, M.; Leone, S. Spontaneous fungal peritonitis: Epidemiology, current evidence and future prospective. World J. Gastroenterol. 2016, 22, 7742–7747. [Google Scholar] [CrossRef] [PubMed]
- Theocharidou, E.; Agarwal, B.; Jeffrey, G.; Jalan, R.; Harrison, D.; Burroughs, A.K.; Kibbler, C.C. Early invasive fungal infections and colonization in patients with cirrhosis admitted to the intensive care unit. Clin. Microbiol. Infect. 2016, 22, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Morrell, M.; Fraser, V.J.; Kollef, M.H. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–3645. [Google Scholar] [CrossRef] [PubMed]
- Vandewoude, K.H.; Blot, S.I.; Benoit, D.; Colardyn, F.; Vogelaers, D. Invasive aspergillosis in critically ill patients: Attributable mortality and excesses in length of icu stay and ventilator dependence. J. Hosp. Infect. 2004, 56, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Verweij, P.E.; Arendrup, M.C.; Arikan-Akdagli, S.; Bille, J.; Donnelly, J.P.; Jensen, H.E.; Lass-Florl, C.; Richardson, M.D.; Akova, M.; et al. Escmid guideline for the diagnosis and management of Candida diseases 2012: Diagnostic procedures. Clin. Microbiol. Infect. 2012, 18, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Cummings, L.A.; Kurosawa, K.; Hoogestraat, D.R.; SenGupta, D.J.; Candra, F.; Doyle, M.; Thielges, S.; Land, T.A.; Rosenthal, C.A.; Hoffman, N.G.; et al. Clinical next generation sequencing outperforms standard microbiological culture for characterizing polymicrobial samples. Clin. Chem. 2016, 62, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Zhang, Y.; Gong, Y.; Sun, R.; Su, L.; Lin, X.; Shen, A.; Zhou, J.; Caiji, Z.; Wang, X.; et al. Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in icu patients. Arch. Med. Res. 2016, 47, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Grumaz, C.; Kirstahler, P.; Sohn, K. The molecular blueprint of a fungus by next-generation sequencing (NGS). Methods Mol. Biol. 2017, 1508, 361–383. [Google Scholar] [PubMed]
- Aquino, V.R.; Nagel, F.; Andreolla, H.F.; de-Paris, F.; Xavier, M.O.; Goldani, L.Z.; Denning, D.W.; Pasqualotto, A.C. The performance of real-time pcr, galactomannan, and fungal culture in the diagnosis of invasive aspergillosis in ventilated patients with chronic obstructive pulmonary disease (COPD). Mycopathologia 2012, 174, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Persat, F.; Ranque, S.; Derouin, F.; Michel-Nguyen, A.; Picot, S.; Sulahian, A. Contribution of the (1,3)-β-d-glucan assay for diagnosis of invasive fungal infections. J. Clin. Microbiol. 2008, 46, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Sulahian, A.; Porcher, R.; Bergeron, A.; Touratier, S.; Raffoux, E.; Menotti, J.; Derouin, F.; Ribaud, P. Use and limits of (1–3)-β-d-glucan assay (fungitell), compared to galactomannan determination (platelia aspergillus), for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 2014, 52, 2328–2333. [Google Scholar] [CrossRef] [PubMed]
- De Pauw, B.; Walsh, T.J.; Donnelly, J.P.; Stevens, D.A.; Edwards, J.E.; Calandra, T.; Pappas, P.G.; Maertens, J.; Lortholary, O.; Kauffman, C.A.; et al. Revised definitions of invasive fungal disease from the european organization for research and treatment of cancer/invasive fungal infections cooperative group and the national institute of allergy and infectious diseases mycoses study group (EORTC/MSG) consensus group. Clin. Infect. Dis. 2008, 46, 1813–1821. [Google Scholar] [PubMed]
- Karageorgopoulos, D.E.; Vouloumanou, E.K.; Ntziora, F.; Michalopoulos, A.; Rafailidis, P.I.; Falagas, M.E. β-d-glucan assay for the diagnosis of invasive fungal infections: A meta-analysis. Clin. Infect. Dis. 2011, 52, 750–770. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, Y.Q.; Guo, Y.L.; Qin, S.M.; Wu, C.; Wang, K. Diagnosis of invasive fungal disease using serum (1→3)-β-d-glucan: A bivariate meta-analysis. Intern. Med. 2011, 50, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.Y.; Wang, S.H.; Liang, S.X.; Jiang, W.X.; Luo, D.D.; Huang, D.H. The screening performance of serum 1,3-β-d-glucan in patients with invasive fungal diseases: A meta-analysis of prospective cohort studies. PLoS ONE 2015, 10, e0131602. [Google Scholar] [CrossRef] [PubMed]
- Digby, J.; Kalbfleisch, J.; Glenn, A.; Larsen, A.; Browder, W.; Williams, D. Serum glucan levels are not specific for presence of fungal infections in intensive care unit patients. Clin. Diagn. Lab. Immunol. 2003, 10, 882–885. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.M.; Lowry, C.M.; Lempitski, S.J.; Kubiak, D.W.; Finkelman, M.A.; Baden, L.R. Reactivity of (1→3)-β-d-glucan assay with commonly used intravenous antimicrobials. Antimicrob. Agents Chemother. 2006, 50, 3450–3453. [Google Scholar] [CrossRef] [PubMed]
- Zedek, D.C.; Miller, M.B. Use of galactomannan enzyme immunoassay for diagnosis of invasive aspergillosis in a tertiary-care center over a 12-month period. J. Clin. Microbiol. 2006, 44, 1601. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W. Invasive aspergillosis. Clin. Infect. Dis. 1998, 26, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Montagna, M.T.; Lovero, G.; Coretti, C.; Martinelli, D.; Delia, M.; de Giglio, O.; Caira, M.; Puntillo, F.; D’Antonio, D.; Venditti, M.; et al. Simiff study: Italian fungal registry of mold infections in hematological and non-hematological patients. Infection 2014, 42, 141–151. [Google Scholar] [CrossRef]
- Meersseman, W.; Lagrou, K.; Maertens, J.; van Wijngaerden, E. Invasive aspergillosis in the intensive care unit. Clin. Infect. Dis. 2007, 45, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Baddley, J.W.; Stephens, J.M.; Ji, X.; Gao, X.; Schlamm, H.T.; Tarallo, M. Aspergillosis in intensive care unit (ICU) patients: Epidemiology and economic outcomes. BMC Infect. Dis. 2013. [Google Scholar] [CrossRef] [PubMed]
- Taccone, F.S.; Van den Abeele, A.M.; Bulpa, P.; Misset, B.; Meersseman, W.; Cardoso, T.; Paiva, J.A.; Blasco-Navalpotro, M.; de Laere, E.; Dimopoulos, G.; et al. Epidemiology of invasive aspergillosis in critically ill patients: Clinical presentation, underlying conditions, and outcomes. Crit. Care 2015, 19, 7. [Google Scholar] [CrossRef] [Green Version]
- Koulenti, D.; Vogelaers, D.; Blot, S. What’s new in invasive pulmonary aspergillosis in the critically ill. Intensive Care Med. 2014, 40, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Hope, W.W.; Walsh, T.J.; Denning, D.W. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 2005, 5, 609–622. [Google Scholar] [CrossRef]
- Leeflang, M.M.; Debets-Ossenkopp, Y.J.; Wang, J.; Visser, C.E.; Scholten, R.J.; Hooft, L.; Bijlmer, H.A.; Reitsma, J.B.; Zhang, M.; Bossuyt, P.M.; et al. Galactomannan detection for invasive aspergillosis in immunocompromised patients. Cochrane Database Syst. Rev. 2015, CD007394. [Google Scholar] [CrossRef] [Green Version]
- Meersseman, W.; Vandecasteele, S.J.; Wilmer, A.; Verbeken, E.; Peetermans, W.E.; van Wijngaerden, E. Invasive aspergillosis in critically ill patients without malignancy. Am. J. Respir. Crit. Care Med. 2004, 170, 621–625. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Ding, L.; Chang, S.; Li, F.; Zhan, Q. Value of consecutive galactomannan determinations for the diagnosis and prognosis of invasive pulmonary aspergillosis in critically ill chronic obstructive pulmonary disease. Med. Mycol. 2011, 49, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Meersseman, W.; Lagrou, K.; Maertens, J.; Wilmer, A.; Hermans, G.; Vanderschueren, S.; Spriet, I.; Verbeken, E.; van Wijngaerden, E. Galactomannan in bronchoalveolar lavage fluid: A tool for diagnosing aspergillosis in intensive care unit patients. Am. J. Respir. Crit. Care Med. 2008, 177, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.; Simon, M.; Katchanov, J.; Wijaya, C.; Rohde, H.; Christner, M.; Laqmani, A.; Wichmann, D.; Fuhrmann, V.; Kluge, S. Does galactomannan testing increase diagnostic accuracy for IPA in the ICU? A prospective observational study. Crit. Care 2016, 20, 139. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Tang, L.; Zhao, S.; Zhao, Z.; Chen, L.; Chen, P.; Huang, Z.; Li, J.; Chen, L.; Fan, X. Systematic review and meta-analysis of detecting galactomannan in bronchoalveolar lavage fluid for diagnosing invasive aspergillosis. PLoS ONE 2012, 7, e43347. [Google Scholar] [CrossRef] [PubMed]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2011, 11, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2004, 4, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Bozzi, A.; Reis, B.S.; Goulart, M.I.; Pereira, M.C.; Pedroso, E.P.; Goes, A.M. Analysis of memory t cells in the human paracoccidioidomycosis before and during chemotherapy treatment. Immunol. Lett. 2007, 114, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Brasch, J. Pathogenesis of tinea. J. Dtsch. Dermatol. Ges 2010, 8, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Muller, U.; Stenzel, W.; Kohler, G.; Werner, C.; Polte, T.; Hansen, G.; Schutze, N.; Straubinger, R.K.; Blessing, M.; McKenzie, A.N.; et al. IL-13 induces disease-promoting type 2 cytokines, alternatively activated macrophages and allergic inflammation during pulmonary infection of mice with cryptococcus neoformans. J. Immunol. 2007, 179, 5367–5377. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, W.A.; Deepe, G.S., Jr. The CCL7-CCL2-CCR2 axis regulates IL-4 production in lungs and fungal immunity. J. Immunol. 2009, 183, 1964–1974. [Google Scholar] [CrossRef] [PubMed]
- Miossec, P.; Korn, T.; Kuchroo, V.K. Interleukin-17 and type 17 helper t cells. N. Engl. J. Med. 2009, 361, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Joosten, L.A.; Kullberg, B.J.; Netea, M.G. Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 2012, 80, 1304–1313. [Google Scholar] [CrossRef]
- Matsuzaki, G.; Umemura, M. Interleukin-17 as an effector molecule of innate and acquired immunity against infections. Microbiol. Immunol. 2007, 51, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Camargo, J.F.; Husain, S. Immune correlates of protection in human invasive aspergillosis. Clin. Infect. Dis. 2014, 59, 569–577. [Google Scholar] [CrossRef]
- Krause, R.; Zollner-Schwetz, I.; Salzer, H.J.; Valentin, T.; Rabensteiner, J.; Pruller, F.; Raggam, R.; Meinitzer, A.; Prattes, J.; Rinner, B.; et al. Elevated levels of interleukin 17a and kynurenine in candidemic patients, compared with levels in noncandidemic patients in the intensive care unit and those in healthy controls. J. Infect. Dis. 2015, 211, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; van de Veerdonk, F.; Smeekens, S.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; Netea, M.G. Candida albicans dampens host defense by downregulating IL-17 production. J. Immunol. 2010, 185, 2450–2457. [Google Scholar] [CrossRef] [PubMed]
- Flierl, M.A.; Rittirsch, D.; Gao, H.; Hoesel, L.M.; Nadeau, B.A.; Day, D.E.; Zetoune, F.S.; Sarma, J.V.; Huber-Lang, M.S.; Ferrara, J.L.; et al. Adverse functions of IL-17a in experimental sepsis. FASEB J. 2008, 22, 2198–2205. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; Puccetti, P. Protective tolerance to fungi: The role of IL-10 and tryptophan catabolism. Trends Microbiol. 2006, 14, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Sanchez, F.; Valenzuela-Mendez, B.; Rodriguez-Gutierrez, J.F.; Estella-Garcia, A.; Gonzalez-Garcia, M.A. New role of biomarkers: Mid-regional pro-adrenomedullin, the biomarker of organ failure. Ann. Transl. Med. 2016, 4, 329. [Google Scholar] [CrossRef]
- Angeletti, S.; Battistoni, F.; Fioravanti, M.; Bernardini, S.; Dicuonzo, G. Procalcitonin and mid-regional pro-adrenomedullin test combination in sepsis diagnosis. Clin. Chem. Lab. Med. 2013, 51, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Christ-Crain, M.; Morgenthaler, N.G.; Struck, J.; Harbarth, S.; Bergmann, A.; Muller, B. Mid-regional pro-adrenomedullin as a prognostic marker in sepsis: An observational study. Crit. Care 2005, 9, R816–824. [Google Scholar] [CrossRef] [PubMed]
- Andaluz-Ojeda, D.; Nguyen, H.B.; Meunier-Beillard, N.; Cicuendez, R.; Quenot, J.P.; Calvo, D.; Dargent, A.; Zarca, E.; Andres, C.; Nogales, L.; et al. Superior accuracy of mid-regional proadrenomedullin for mortality prediction in sepsis with varying levels of illness severity. Ann. Intensive Care 2017, 7, 15. [Google Scholar] [CrossRef]
- Zudaire, E.; Portal-Nunez, S.; Cuttitta, F. The central role of adrenomedullin in host defense. J. Leukoc. Biol. 2006, 80, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Hinson, J.P.; Kapas, S.; Smith, D.M. Adrenomedullin, a multifunctional regulatory peptide. Endocr. Rev. 2000, 21, 138–167. [Google Scholar] [CrossRef]
- Allaker, R.P.; Grosvenor, P.W.; McAnerney, D.C.; Sheehan, B.E.; Srikanta, B.H.; Pell, K.; Kapas, S. Mechanisms of adrenomedullin antimicrobial action. Peptides 2006, 27, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Brell, B.; Temmesfeld-Wollbruck, B.; Altzschner, I.; Frisch, E.; Schmeck, B.; Hocke, A.C.; Suttorp, N.; Hippenstiel, S. Adrenomedullin reduces staphylococcus aureus alpha-toxin-induced rat ileum microcirculatory damage. Crit. Care Med. 2005, 33, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Allaker, R.P.; Zihni, C.; Kapas, S. An investigation into the antimicrobial effects of adrenomedullin on members of the skin, oral, respiratory tract and gut microflora. FEMS Immunol. Med. Microbiol. 1999, 23, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Hiemenz, J.W.; Seibel, N.L.; Perfect, J.R.; Horwith, G.; Lee, L.; Silber, J.L.; DiNubile, M.J.; Reboli, A.; Bow, E.; et al. Amphotericin B lipid complex for invasive fungal infections: Analysis of safety and efficacy in 556 cases. Clin. Infect. Dis. 1998, 26, 1383–1396. [Google Scholar] [CrossRef]
- Kuhbacher, A.; Henkel, H.; Stevens, P.; Grumaz, C.; Finkelmeier, D.; Burger-Kentischer, A.; Sohn, K.; Rupp, S. Dermal fibroblasts play a central role in skin model protection against C. Albicans invasion. J. Infect. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock: 2012. Crit. Care Med. 2013, 41, 580–637. [Google Scholar] [CrossRef] [PubMed]
- Rivers, E.; Nguyen, B.; Havstad, S.; Ressler, J.; Muzzin, A.; Knoblich, B.; Peterson, E.; Tomlanovich, M. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N. Engl. J. Med. 2001, 345, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Weigand, M.A.; Bardenheuer, H.J.; Bottiger, B.W. Clinical management of patients with sepsis. Anaesthesist 2003, 52, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Gumbinger, C.; Hug, A.; Murle, B.; Berger, B.; Zorn, M.; Becker, K.P.; Zimmermann, S.; Dalpke, A.H.; Veltkamp, R. Early blood-based microbiological testing is ineffective in severe stroke patients. J. Neurol. Sci. 2013, 325, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Mischnik, A.; Mieth, M.; Busch, C.J.; Hofer, S.; Zimmermann, S. First evaluation of automated specimen inoculation for wound swab samples by use of the previ isola system compared to manual inoculation in a routine laboratory: Finding a cost-effective and accurate approach. J. Clin. Microbiol. 2012, 50, 2732–2736. [Google Scholar] [CrossRef] [PubMed]
- Werle, E.; Kappe, R.; Fiehn, W.; Sonntag, H.G. Detection of anti-Candida antibodies of the classes IgM, IgG and IgA using enzyme immunoassay in sequential serum samples of hospitalized patients. Mycoses 1994, 37 (Suppl. 1), 71–78. [Google Scholar] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlazeck, F.J.; Rescheneder, P.; von Haeseler, A. Nextgenmap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]








| Parameter | Unit | All Patients (n = 50) | Without Fungal Isolates (n = 17) | With Fungal Isolates (n = 33) | p for Patients without Fungal Isolates vs. Patients with Fungal Isolates |
|---|---|---|---|---|---|
| Gender male | 38 (76) | 11 (64.7) | 27 (81.8) | 0.160 | |
| Age | (years) | 66 (61–75) | 71 (64–80) | 66 (59–74) | 0.117 |
| BMI | (kg/m2) | 27.2 (24.4–30.9) | 27.2 (25.7–34.9) | 26.9 (23.1–30.9) | 0.401 |
| Postoperative peritonitis | 31 | 9 (52.9) | 22 (66.7) | 0.206 | |
| initial operation | |||||
| Kidney | 2 (4) | 0 (0) | 2 (6.1) | 0.431 | |
| Liver | 11 (22) | 1 (2.1) | 10 (30.3) | 0.047 * | |
| Pancreas | 2 (10) | 1 (5.9) | 1 (3.0) | 0.569 | |
| GIT | 38 (76) | 14 (82.4) | 24 (72.7) | 0.350 | |
| VAS | 3 (6) | 2 (11.8) | 1 (3.0) | 0.264 | |
| Others | 12 (24) | 3 (17.6) | 9 (27.3) | 0.350 | |
| ≥48 h after hospital admission | 25 (50) | 7 (41.2) | 18 (54.5) | 0.276 | |
| NYHA 0-I | 41 (82) | 13 (76.4) | 28 (84.8) | 0.358 | |
| Diabetes mellitus | 17 (34) | 5 (29.4) | 12 (36.3) | 0.434 | |
| Arterial hypertension | 34 (68) | 12 (70.6) | 22 (66.7) | 0.520 | |
| Coronary heart disease | 8 (16) | 5 (29.4) | 3 (9.1) | 0.076 | |
| Chronic obstructive lung disease | 10 (20) | 5 (29.4) | 5 (15.2) | 0.204 | |
| Renal insufficiency | 7 (14) | 1 (5.9) | 6 (18.2) | 0.231 | |
| Renal replacement therapy | 15 (30) | 2 (11.8) | 13 (39.4) | 0.041 * | |
| Liver cirrhosis | 13 (26) | 3 (17.6) | 10 (30.3) | 0.270 | |
| Oncological disease | 33 (66) | 11 (64.7) | 22 (66.7) | 0.566 | |
| APACHE II # | 30 (28–35) | 32 (28–36) | 30 (28–34) | 0.491 | |
| SOFA # | 11 (10–14) | 11 (10–14) | 11 (10–14) | 0.959 | |
| SAPS II # | 65 (49–75) | 72 (48–75) | 65 (51–72) | 0.467 | |
| Candida colonization | 22 (44) | 0 (0) | 22 (66.7) | --- | |
| Candida infection | 10 (20) | 0 (0) | 10 (30.3) | --- | |
| Candidemia | 3 (6) | 0 (0) | 3 (9.1) | --- | |
| Aspergillus spp. | 1 (3) | 0 (0) | 1 (3.0) | --- | |
| Candida-Score | 4 (4–4) | 4 (4–4) | 4 (4–4) | 0.080 | |
| Ventilation duration | (h) | 145.5 (67.3–450) | 89 (46–145) | 181 (77–682) | 0.015 * |
| ICU length of stay | (days) | 19.5 (12–44) | 12 (3–17) | 24 (15–46) | 0.002 ** |
| Hospital length of stay | (days) | 44 (23.3–68.5) | 24 (12–40) | 51 (39–78) | 0.007 ** |
| Tracheotomy | 14 (28) | 2 (11.8) | 12 (36.3) | 0.063 | |
| Anastomosis leakage | 24 (48) | 7 (41.2) | 17 (51.5) | 0.347 | |
| Fascia dehiscence | 12 (24) | 2 (11.8) | 10 (30.3) | 0.134 | |
| 90 day mortality | 17 (34) | 8 (47.1) | 9 (27.3) | 0.175 | |
| 28 day mortality | 11 (22) | 7 (41.2) | 4 (12.1) | 0.025 * |
| Parameter | Unit | Fungal Colonization (n = 22) | Fungal Infection (n = 11) | p for Patients with Fungal Colonization vs. Patients with Fungal Infection |
|---|---|---|---|---|
| Male | 17 (77.3) | 10 (90.1) | 0.329 | |
| Age | (years) | 66 (61–74) | 65 (58–74) | 0.355 |
| BMI | (kg/m2) | 25.3 (21.6–30.8) | 27.4 (26–30.5) | 0.925 |
| Postoperative peritonitis | 14 (63.6) | 8 (72.7) | 0.454 | |
| Initial operation | ||||
| Kidney | 1 (4.5) | 1 (9.1) | 0.563 | |
| Liver | 3 (13.6) | 7 (63.6) | 0.006 ** | |
| Pancreas | 1 (4.5) | 0 (0) | 0.667 | |
| GIT | 16 (72.7) | 8 (72.7) | 0.653 | |
| VAS | 0 (0) | 1 (9.1) | 0.333 | |
| Others | 5 (22.7) | 4 (36.4) | 0.333 | |
| ≥48 h after hospital admission | 15 (68.2) | 3 (27.3) | 0.031 * | |
| NYHA 0-I | 17 (77.3) | 11 (100) | 0.111 | |
| Diabetes mellitus | 8 (36.4) | 4 (36.4) | 0.653 | |
| Arterial hypertension | 15 (68.2) | 7 (63.6) | 0.546 | |
| Coronary heart disease | 2 (9.1) | 1 (9.1) | 0.748 | |
| Chronic obstructive lung disease | 5 (22.7) | 0 (0) | 0.111 | |
| Renal insufficiency | 5 (22.7) | 1 (9.1) | 0.329 | |
| Renal replacement therapy | 8 (36.4) | 5 (45.5) | 0.446 | |
| Liver cirrhosis | 3 (13.6) | 7 (63.6) | 0.006 ** | |
| Oncological disease | 14 (63.6) | 8 (72.7) | 0.454 | |
| APACHE II # | 30 (29–34) | 29 (28–33) | 0.396 | |
| SOFA # | 11 (10–13) | 14 (11–15) | 0.044 * | |
| SAPS II # | 61 (44–72) | 68 (57–77) | 0.336 | |
| Candida-Score | 4 (4–4) | 4 (4–4) | 1.0 | |
| Ventilation duration | (h) | 148.5 (74–239.3) | 600 (424.5–944) | 0.040 * |
| Tracheotomy | 2 (9.1) | 8 (72.7) | 0.002 ** | |
| Fascia dehiscence | 3 (13.6) | 7 (63.6) | 0.006 ** | |
| Anastomosis leakage | 11 (50) | 6 (54.5) | 0.549 | |
| ICU length of stay | (days) | 21 (13.5–43.5) | 38 (25.5–64) | 0.082 |
| Hospital length of stay | (days) | 50 (34.5–68.5) | 53 (47.5–88) | 0.418 |
| 90 day mortality | 4 (18.2) | 5 (45.5) | 0.120 | |
| 28 day mortality | 3 (13.6) | 1 (9.1) | 0.593 |
| Parameter | Unit | Without Fungal Infection (n = 39) | Fungal Infection (n = 11) | p for Patients without Fungal Infection vs. Patients with Fungal Infection |
|---|---|---|---|---|
| Gender male | 28 (71.8) | 10 (90.1) | 0.184 | |
| Age | (years) | 66 (63–76) | 65 (58–74) | 0.460 |
| BMI | (kg/m2) | 27.2 (23.2–33.2) | 27.4 (26–30.5) | 0.582 |
| Postoperative peritonitis | 23 (58.9) | 8 (72.7) | 0.322 | |
| initial operation | ||||
| Kidney | 1 (2.6) | 1 (9.1) | 0.395 | |
| Liver | 4 (10.3) | 7 (63.6) | 0.001 *** | |
| Pancreas | 2 (5.1) | 0 (0) | 0.605 | |
| GIT | 30 (76.9) | 8 (72.7) | 0.528 | |
| VAS | 2 (5.1) | 1 (9.1) | 0.534 | |
| Others | 8 (20.5) | 4 (36.4) | 0.240 | |
| ≥48 h after hospital admission | 22 (56.4) | 3 (27.3) | 0.085 | |
| NYHA 0-I | 30 (76.9) | 11 (100) | 0.115 | |
| Diabetes mellitus | 13 (33.3) | 4 (36.4) | 0.560 | |
| Arterial hypertension | 27 (69.2) | 7 (63.6) | 0.495 | |
| Coronary heart disease | 7 (17.9) | 1 (9.1) | 0.430 | |
| Chronic obstructive lung disease | 10 (25.6) | 0 (0) | 0.062 | |
| Renal insufficiency | 6 (15.4) | 1 (9.1) | 0.604 | |
| Renal replacement therapy | 10 (25.6) | 5 (45.5) | 0.184 | |
| Liver cirrhosis | 6 (15.4) | 7 (63.6) | 0.003 ** | |
| Oncological disease | 25 (64.1) | 8 (72.7) | 0.440 | |
| APACHE II # | 31 (28–36) | 29 (28–33) | 0.335 | |
| SOFA # | 11 (10–13) | 14 (11–15) | 0.081 | |
| SAPS II # | 62 (47–74) | 68 (57–77) | 0.519 | |
| Candida-Score | 4 (4–4) | 4 (4–4) | 0.881 | |
| Candida colonization | 22 (56.4) | 0 (0%) | --- | |
| Candida infection | 0 (0%) | 10 (90.1) | --- | |
| Candidemia | 0 (0%) | 3 (27.3) | --- | |
| Aspergillus spp. | 0 (0%) | 1 (9.1) | --- | |
| Ventilation duration | (h) | 120 (60–200) | 600 (424.5–944) | 0.007 ** |
| ICU length of stay | (days) | 16 (10–28.5) | 38 (25.5–64) | 0.008 ** |
| Hospital length of stay | (days) | 40 (21.5–63) | 53 (47.5–88) | 0.075 |
| Tracheotomy | 4 (10.3) | 8 (72.7) | 0.001 *** | |
| Anastomosis leakage | 18 (46.2) | 6 (54.5) | 0.440 | |
| Fascia dehiscence | 13 (33.3) | 7 (63.6) | 0.002 ** | |
| 90 day mortality | 12 (30.8) | 5 (45.5) | 0.293 | |
| 28 day mortality | 10 (25.6) | 1 (9.1) | 0.232 |
| First-Line Antifungal Therapy | n = 21 (%) |
|---|---|
| Fluconazole | 7 (33.3) |
| Caspofungin | 13 (61.9) |
| Liposomal amphotericin B | 1 (4.8) |
| Used as empiric therapy | 7 (33.3) |
| Fluconazole | 1 (14.3) |
| Caspofungin | 6 (85.7) |
| Change in antifungal therapy | 5 (23.9) |
| Second-Line Antifungal Therapy | n = 5 (%) |
| Caspofungin | 4 (80.0) |
| Fluconazole | 1 (20.0) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Decker, S.O.; Sigl, A.; Grumaz, C.; Stevens, P.; Vainshtein, Y.; Zimmermann, S.; Weigand, M.A.; Hofer, S.; Sohn, K.; Brenner, T. Immune-Response Patterns and Next Generation Sequencing Diagnostics for the Detection of Mycoses in Patients with Septic Shock—Results of a Combined Clinical and Experimental Investigation. Int. J. Mol. Sci. 2017, 18, 1796. https://doi.org/10.3390/ijms18081796
Decker SO, Sigl A, Grumaz C, Stevens P, Vainshtein Y, Zimmermann S, Weigand MA, Hofer S, Sohn K, Brenner T. Immune-Response Patterns and Next Generation Sequencing Diagnostics for the Detection of Mycoses in Patients with Septic Shock—Results of a Combined Clinical and Experimental Investigation. International Journal of Molecular Sciences. 2017; 18(8):1796. https://doi.org/10.3390/ijms18081796
Chicago/Turabian StyleDecker, Sebastian O., Annette Sigl, Christian Grumaz, Philip Stevens, Yevhen Vainshtein, Stefan Zimmermann, Markus A. Weigand, Stefan Hofer, Kai Sohn, and Thorsten Brenner. 2017. "Immune-Response Patterns and Next Generation Sequencing Diagnostics for the Detection of Mycoses in Patients with Septic Shock—Results of a Combined Clinical and Experimental Investigation" International Journal of Molecular Sciences 18, no. 8: 1796. https://doi.org/10.3390/ijms18081796