MicroRNA–mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers
Abstract
:1. Introduction:
1.1. Placenta and Pregnancy Complications
1.2. MicroRNAs as Biomarkers of Pregnancy Complications
2. Identification of miRNA–mRNA Networks Linked to Placental Development
2.1. MicroRNA Inclusion Criteria
2.2. Identification of miRNA Target Genes
2.3. Downstream Functional Analysis
3. Findings and Discussion
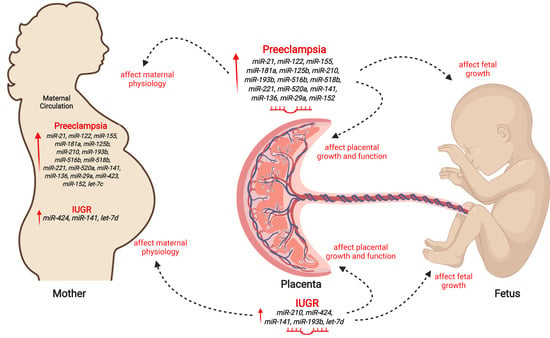
3.1. MicroRNAs in PE and IUGR
3.2. MicroRNA–mRNA Networks in PE
3.3. MicroRNA–mRNA Networks in IUGR
3.4. Pathways and Biological Processes Affected by Potential Biomarkers of PE
3.5. Pathways and Biological Processes Affected by Potential Biomarkers of IUGR
3.6. MicroRNA-210
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gude, N.M.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L.; Thornburg, K.L. Placental origins of chronic disease. Physiol. Rev. 2016, 96, 1509–1565. [Google Scholar] [CrossRef] [PubMed]
- Turco, M.Y.; Moffett, A. Development of the human placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. The Role of LIN28-let-7-ARID3B Pathway in Placental Development. Int. J. Mol. Sci. 2020, 21, 3637. [Google Scholar] [CrossRef]
- Reed, M.D.; Mattison, D.R. Clinical Pharmacology During Pregnancy; Elsevier: London, UK, 2013; p. 5. [Google Scholar]
- Skinner, M.K. Encyclopedia of Reproduction; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Georgiades, P.; Ferguson-Smith, A.; Burton, G. Comparative developmental anatomy of the murine and human definitive placentae. Placenta 2002, 23, 3–19. [Google Scholar] [CrossRef]
- Huppertz, B.; Kaufmann, P. Trophoblast turnover in health and disease. Fetal Matern. Med. Rev. 2002, 13, 103. [Google Scholar] [CrossRef]
- Yang, H.; Ma, Q.; Wang, Y.; Tang, Z. Clinical application of exosomes and circulating microRNAs in the diagnosis of pregnancy complications and foetal abnormalities. J. Transl. Med. 2020, 18, 32. [Google Scholar] [CrossRef]
- Awamleh, Z.; Gloor, G.B.; Han, V.K. Placental microRNAs in pregnancies with early onset intrauterine growth restriction and preeclampsia: Potential impact on gene expression and pathophysiology. BMC Med. Genom. 2019, 12, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armengaud, J.; Yzydorczyk, C.; Siddeek, B.; Peyter, A.; Simeoni, U. Intrauterine growth restriction: Clinical consequences on health and disease at adulthood. Reprod. Toxicol. 2020, 99, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Vayssière, C.; Sentilhes, L.; Ego, A.; Bernard, C.; Cambourieu, D.; Flamant, C.; Gascoin, G.; Gaudineau, A.; Grangé, G.; Houfflin-Debarge, V. Fetal growth restriction and intra-uterine growth restriction: Guidelines for clinical practice from the French College of Gynaecologists and Obstetricians. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015, 193, 10–18. [Google Scholar] [CrossRef]
- Easter, S.R.; Eckert, L.O.; Boghossian, N.; Spencer, R.; Oteng-Ntim, E.; Ioannou, C.; Patwardhan, M.; Harrison, M.S.; Khalil, A.; Gravett, M. Fetal growth restriction: Case definition & guidelines for data collection, analysis, and presentation of immunization safety data. Vaccine 2017, 35, 6546. [Google Scholar]
- Barker, D.J. Adult consequences of fetal growth restriction. Clin. Obstet. Gynecol. 2006, 49, 270–283. [Google Scholar] [CrossRef]
- Baschat, A.A.; Hecher, K. Fetal Growth Restriction Due to Placental Disease. In Seminars in Perinatology; Elsevier: Amsterdam, The Netherlands, 2004; pp. 67–80. [Google Scholar]
- Lager, S.; Powell, T.L. Regulation of nutrient transport across the placenta. J. Pregnancy 2012, 2012, 179827. [Google Scholar] [CrossRef] [Green Version]
- Barut, F.; Barut, A.; Gun, B.D.; Kandemir, N.O.; Harma, M.I.; Harma, M.; Aktunc, E.; Ozdamar, S.O. Intrauterine growth restriction and placental angiogenesis. Diagn. Pathol. 2010, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brett, K.E.; Ferraro, Z.M.; Yockell-Lelievre, J.; Gruslin, A.; Adamo, K.B. Maternal–fetal nutrient transport in pregnancy pathologies: The role of the placenta. Int. J. Mol. Sci. 2014, 15, 16153–16185. [Google Scholar] [CrossRef] [Green Version]
- Jakó, M.; Surányi, A.; Kaizer, L.; Németh, G.; Bártfai, G. Maternal Hematological Parameters and Placental and Umbilical Cord Histopathology in Intrauterine Growth Restriction. Med. Princ. Pract. 2019, 28, 101–108. [Google Scholar] [CrossRef]
- Gathiram, P.; Moodley, J. Pre-eclampsia: Its pathogenesis and pathophysiolgy. Cardiovasc. J. Afr. 2016, 27, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, G.J.; Redman, C.W.; Roberts, J.M.; Moffett, A. Pre-eclampsia: Pathophysiology and clinical implications. BMJ 2019, 366, 12381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornakova, A.; Kolkova, Z.; Holubekova, V.; Loderer, D.; Lasabova, Z.; Biringer, K.; Halasova, E. Diagnostic Potential of MicroRNAs as Biomarkers in the Detection of Preeclampsia. Genet. Test. Mol. Biomark. 2020, 24, 321–327. [Google Scholar] [CrossRef]
- Roberts, J.M.; Escudero, C. The placenta in preeclampsia. Pregnancy Hypertens. Int. J. Women Cardiovasc. Health 2012, 2, 72–83. [Google Scholar]
- Figueras, F.; Caradeux, J.; Crispi, F.; Eixarch, E.; Peguero, A.; Gratacos, E. Diagnosis and surveillance of late-onset fetal growth restriction. Am. J. Obstet. Gynecol. 2018, 218, S790–S802.e1. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Kang, Y.; Lee, E.M.; Jung, Y.M.; Hong, S.; Park, S.J.; Park, C.-W.; Norwitz, E.R.; Lee, D.Y.; Park, J.S. Metabolomic biomarkers in midtrimester maternal plasma can accurately predict the development of preeclampsia. Sci. Rep. 2020, 10, 16142. [Google Scholar] [CrossRef]
- Tsochandaridis, M.; Nasca, L.; Toga, C.; Levy-Mozziconacci, A. Circulating microRNAs as clinical biomarkers in the predictions of pregnancy complications. Biomed. Res. Int. 2015, 2015, 294954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-W.; Xie, C.; Yang, J.-R.; Li, J.-H.; Yang, J.-H.; Zheng, L. MtiBase: A database for decoding microRNA target sites located within CDS and 5′ UTR regions from CLIP-Seq and expression profile datasets. Database 2015, 2015, bav102. [Google Scholar] [CrossRef] [Green Version]
- Addo, K.A.; Palakodety, N.; Hartwell, H.J.; Tingare, A.; Fry, R.C. Placenta microRNAs: Responders to environmental chemicals and mediators of pathophysiology of the human placenta. Toxicol. Rep. 2020, 7, 1046–1056. [Google Scholar] [CrossRef]
- Hayder, H.; O’Brien, J.; Nadeem, U.; Peng, C. MicroRNAs: Crucial regulators of placental development. Reproduction 2018, 155, R259–R271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotlabova, K.; Doucha, J.; Hromadnikova, I. Placental-specific microRNA in maternal circulation–identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 2011, 89, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.; Pan, W.; Gawad, C.; Fan, H.C.; Kerchner, G.A.; Wyss-Coray, T.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 7361–7366. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, J.; Jäck, H.-M. Serum microRNAs as powerful cancer biomarkers. Biochim. Et Biophys. Acta BBA Rev. Cancer 2010, 1806, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Wang, Y.; Quan, Y.; Wang, Z.; Liu, Y.; Ding, Z. Maternal obesity alters C19MC microRNAs expression profile in fetal umbilical cord blood. Nutr. Metab. 2020, 17, 52. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.C.; Mackie, F.L.; Lean, S.C.; Greenwood, S.L.; Heazell, A.E.; Forbes, K.; Jones, R.L. Placental dysfunction is associated with altered microRNA expression in pregnant women with low folate status. Mol. Nutr. Food Res. 2017, 61, 1600646. [Google Scholar] [CrossRef] [Green Version]
- Tsamou, M.; Martens, D.S.; Winckelmans, E.; Madhloum, N.; Cox, B.; Gyselaers, W.; Nawrot, T.S.; Vrijens, K. Mother’s Pre-pregnancy BMI and Placental Candidate miRNAs: Findings from the ENVIR ON AGE Birth Cohort. Sci. Rep. 2017, 7, 5548. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.-S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, K.; Miura, S.; Yamasaki, K.; Higashijima, A.; Kinoshita, A.; Yoshiura, K.-i.; Masuzaki, H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010, 56, 1767–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, M.; Kolluru, G.K.; Ahmed, A. Small molecule, big prospects: MicroRNA in pregnancy and its complications. J. Pregnancy 2017, 2017, 6972732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Lu, J.; Wang, S.; Li, H.; Ge, Q.; Lu, Z. Application of next-generation sequencing technology to profile the circulating microRNAs in the serum of preeclampsia versus normal pregnant women. Clin. Chim. Acta 2011, 412, 2167–2173. [Google Scholar] [CrossRef]
- Sibai, B.M. Preeclampsia as a Cause of Preterm and Late Preterm (Near-Term) Births. In Seminars in Perinatology; Elsevier: Amsterdam, The Netherlands, 2006; pp. 16–19. [Google Scholar]
- Azar, C.; Valentine, M.; Trausch-Azar, J.; Druley, T.; Nelson, D.M.; Schwartz, A.L. RNA-Seq identifies genes whose proteins are transformative in the differentiation of cytotrophoblast to syncytiotrophoblast, in human primary villous and BeWo trophoblasts. Sci. Rep. 2018, 8, 5142. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Biró, O.; Fóthi, Á.; Alasztics, B.; Nagy, B.; Orbán, T.I.; Rigó, J., Jr. Circulating exosomal and Argonaute-bound microRNAs in preeclampsia. Gene 2019, 692, 138–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, L.; Liu, Z.; Wei, M.; Chen, Y.; Yang, X.; Chen, L.; Xiao, X. MiR-210 and miR-155 as potential diagnostic markers for pre-eclampsia pregnancies. Medicine 2017, 96, e7515. [Google Scholar] [CrossRef] [PubMed]
- Gunel, T.; Zeybek, Y.; Akçakaya, P.; Kalelioglu, I.; Benian, A.; Ermis, H.; Aydinli, K. Serum microRNA expression in pregnancies with preeclampsia. Genet. Mol. Res. 2011, 10, 4034–4040. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jairajpuri, D.S.; Malalla, Z.H.; Mahmood, N.; Almawi, W.Y. Circulating microRNA expression as predictor of preeclampsia and its severity. Gene 2017, 627, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Long, A.; Tan, L.; Hong, M.; Wu, J.; Cai, L.; Li, Q. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 2017, 51, 70–75. [Google Scholar] [CrossRef]
- Li, H.; Ge, Q.; Guo, L.; Lu, Z. Maternal plasma miRNAs expression in preeclamptic pregnancies. BioMed Res. Int. 2013, 2013, 970265. [Google Scholar] [CrossRef]
- Li, Q.; Long, A.; Jiang, L.; Cai, L.; Xie, L.; Gu, J.A.; Chen, X.; Tan, L. Quantification of preeclampsia-related microRNAs in maternal serum. Biomed. Rep. 2015, 3, 792–796. [Google Scholar] [CrossRef]
- Luque, A.; Farwati, A.; Crovetto, F.; Crispi, F.; Figueras, F.; Gratacós, E.; Aran, J.M. Usefulness of circulating microRNAs for the prediction of early preeclampsia at first-trimester of pregnancy. Sci. Rep. 2014, 4, 4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Fierro, M.L.; Carrillo-Arriaga, J.G.; Luevano, M.; Lugo-Trampe, A.; Delgado-Enciso, I.; Rodriguez-Sanchez, I.P.; Garza-Veloz, I. Serum levels of miR-628-3p and miR-628-5p during the early pregnancy are increased in women who subsequently develop preeclampsia. Pregnancy Hypertens. 2019, 16, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fierro, M.L.; Garza-Veloz, I.; Gutierrez-Arteaga, C.; Delgado-Enciso, I.; Barbosa-Cisneros, O.Y.; Flores-Morales, V.; Hernandez-Delgadillo, G.P.; Rocha-Pizaña, M.R.; Rodriguez-Sanchez, I.P.; Badillo-Almaraz, J.I. Circulating levels of specific members of chromosome 19 microRNA cluster are associated with preeclampsia development. Arch. Gynecol. Obstet. 2018, 297, 365–371. [Google Scholar] [CrossRef]
- Miura, K.; Higashijima, A.; Murakami, Y.; Tsukamoto, O.; Hasegawa, Y.; Abe, S.; Fuchi, N.; Miura, S.; Kaneuchi, M.; Masuzaki, H. Circulating chromosome 19 miRNA cluster microRNAs in pregnant women with severe pre-eclampsia. J. Obstet. Gynaecol. Res. 2015, 41, 1526–1532. [Google Scholar] [CrossRef] [PubMed]
- Motawi, T.M.; Sabry, D.; Maurice, N.W.; Rizk, S.M. Role of mesenchymal stem cells exosomes derived microRNAs; miR-136, miR-494 and miR-495 in pre-eclampsia diagnosis and evaluation. Arch. Biochem. Biophys. 2018, 659, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Pineles, B.L.; Romero, R.; Montenegro, D.; Tarca, A.L.; Han, Y.M.; Kim, Y.M.; Draghici, S.; Espinoza, J.; Kusanovic, J.P.; Mittal, P. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am. J. Obstet. Gynecol. 2007, 196, 261.e1–261.e6. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Tang, Y.; Yang, N.; Wei, X.; Wu, J. Potential role of circulating microRNAs as a biomarker for unexplained recurrent spontaneous abortion. Fertil. Steril. 2016, 105, 1247–1254.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salomon, C.; Guanzon, D.; Scholz-Romero, K.; Longo, S.; Correa, P.; Illanes, S.E.; Rice, G.E. Placental exosomes as early biomarker of preeclampsia: Potential role of exosomal microRNAs across gestation. J. Clin. Endocrinol. Metab. 2017, 102, 3182–3194. [Google Scholar] [CrossRef]
- Stubert, J.; Koczan, D.; Richter, D.-U.; Dieterich, M.; Ziems, B.; Thiesen, H.-J.; Gerber, B.; Reimer, T. miRNA expression profiles determined in maternal sera of patients with HELLP syndrome. Hypertens. Pregnancy 2014, 33, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.V.; Gusar, V.A.; Kan, N.E.; Prozorovskaya, K.N.; Karapetyan, A.O.; Bayev, O.R.; Chagovets, V.V.; Kliver, S.F.; Iakovishina, D.Y.; Frankevich, V.E. Identification of potential early biomarkers of preeclampsia. Placenta 2018, 61, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Ura, B.; Feriotto, G.; Monasta, L.; Bilel, S.; Zweyer, M.; Celeghini, C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan. J. Obstet. Gynecol. 2014, 53, 232–234. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Song, W.-Y.; Xie, Y.; Hu, L.-L.; Hou, X.-M.; Wang, R.; Gao, Y.; Zhang, J.-N.; Zhang, L.; Li, W.-W. miR-181a-5p suppresses invasion and migration of HTR-8/SVneo cells by directly targeting IGF2BP2. Cell Death Dis. 2018, 9, 16. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Lin, H.; Qi, J.; Zhu, C.; Gao, Z.; Wang, H. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction 2012, 143, 389–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Zhao, Y.; Liu, M.; Wang, Y.; Wang, H.; Li, Y.-X.; Zhu, X.; Yao, Y.; Wang, H.; Qiao, J. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 2014, 63, 1276–1284. [Google Scholar] [CrossRef]
- Yoffe, L.; Gilam, A.; Yaron, O.; Polsky, A.; Farberov, L.; Syngelaki, A.; Nicolaides, K.; Hod, M.; Shomron, N. Early detection of preeclampsia using circulating small non-coding RNA. Sci. Rep. 2018, 8, 3401. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.; Marques, T.; Pereira, R.; Sandrim, V. Reduced circulating miR-196b levels is associated with preeclampsia. Pregnancy Hypertens. Int. J. Women Cardiovasc. Health 2014, 4, 11–13. [Google Scholar] [CrossRef]
- Truong, G.; Guanzon, D.; Kinhal, V.; Elfeky, O.; Lai, A.; Longo, S.; Nuzhat, Z.; Palma, C.; Scholz-Romero, K.; Menon, R. Oxygen tension regulates the miRNA profile and bioactivity of exosomes released from extravillous trophoblast cells–liquid biopsies for monitoring complications of pregnancy. PLoS ONE 2017, 12, e0174514. [Google Scholar]
- Chen, J.; Zhao, L.; Wang, D.; Xu, Y.; Gao, H.; Tan, W.; Wang, C. Contribution of regulatory T cells to immune tolerance and association of microRNA-210 and Foxp3 in preeclampsia. Mol. Med. Rep. 2019, 19, 1150–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.-Y.; Yun, J.; Lee, O.-J.; Han, H.-S.; Yeo, M.-K.; Lee, M.-A.; Suh, K.-S. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta 2013, 34, 799–804. [Google Scholar] [CrossRef]
- Dai, X.; Cai, Y. Down-regulation of microRNA let-7d inhibits the proliferation and invasion of trophoblast cells in preeclampsia. J. Cell. Biochem. 2018, 119, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Enquobahrie, D.A.; Abetew, D.F.; Sorensen, T.K.; Willoughby, D.; Chidambaram, K.; Williams, M.A. Placental microRNA expression in pregnancies complicated by preeclampsia. Am. J. Obstet. Gynecol. 2011, 204, e12–e178.e21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Xiao, Y.; Ren, C.; Guo, Y.; Qu, D.; Zhang, J.; Zhu, Y. Up-regulation of miR-517-5p inhibits ERK/MMP-2 pathway: Potential role in preeclampsia. Eur. Rev. Med. Pharm. Sci. 2018, 22, 6599–6608. [Google Scholar]
- Gao, T.; Deng, M.; Wang, Q. Mi RNA-320a inhibits trophoblast cell invasion by targeting estrogen-related receptor-gamma. J. Obstet. Gynaecol. Res. 2018, 44, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, H.; Wei, J. MiR-4421 regulates the progression of preeclampsia by regulating CYP11B2. Eur. Rev. Med. Pharm. Sci. 2018, 22, 1533–1540. [Google Scholar]
- Hu, Y.; Li, P.; Hao, S.; Liu, L.; Zhao, J.; Hou, Y. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin. Chem. Lab. Med. 2009, 47, 923–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, L.; Zhang, L.; Li, Y.; Guo, L.; Cao, N.; Bai, Z.; Song, Y.; Xu, Z.; Zhang, J.; Liu, C. MiR-136 contributes to pre-eclampsia through its effects on apoptosis and angiogenesis of mesenchymal stem cells. Placenta 2017, 50, 102–109. [Google Scholar] [CrossRef]
- Lasabová, Z.; Vazan, M.; Zibolenova, J.; Svecova, I. Overexpression of miR-21 and miR-122 in preeclamptic placentas. Neuroendocr. Lett. 2015, 36, 695–699. [Google Scholar]
- Li, Q.; Pan, Z.; Wang, X.; Gao, Z.; Ren, C.; Yang, W. miR-125b-1-3p inhibits trophoblast cell invasion by targeting sphingosine-1-phosphate receptor 1 in preeclampsia. Biochem. Biophys. Res. Commun. 2014, 453, 57–63. [Google Scholar] [CrossRef]
- Lykoudi, A.; Kolialexi, A.; Lambrou, G.I.; Braoudaki, M.; Siristatidis, C.; Papaioanou, G.K.; Tzetis, M.; Mavrou, A.; Papantoniou, N. Dysregulated placental microRNAs in Early and Late onset Preeclampsia. Placenta 2018, 61, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Niu, Z.-r.; Han, T.; Sun, X.-l.; Luan, L.-x.; Gou, W.-l.; Zhu, X.-m. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am. J. Obstet. Gynecol. 2018, 218, 249.e1–249.e12. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Prieto, S.; Chaiwangyen, W.; Herrmann, J.; Groten, T.; Schleussner, E.; Markert, U.R.; Morales-Prieto, D.M. MicroRNA-141 is upregulated in preeclamptic placentae and regulates trophoblast invasion and intercellular communication. Transl. Res. 2016, 172, 61–72. [Google Scholar] [CrossRef]
- Svecova, I.; Vazan, M.; Zubor, P.; Danko, J.; Lasabova, Z. P40. MIR-21 and mir-221 overexpression in placental tissue of preeclamptic patients. Pregnancy Hypertens. Int. J. Women Cardiovasc. Health 2015, 5, 245. [Google Scholar] [CrossRef]
- Wang, W.; Feng, L.; Zhang, H.; Hachy, S.; Satohisa, S.; Laurent, L.C.; Parast, M.; Zheng, J.; Chen, D.-b. Preeclampsia up-regulates angiogenesis-associated microRNA (ie. miR-17,-20a, and-20b) that target ephrin-B2 and EPHB4 in human placenta. J. Clin. Endocrinol. Metab. 2012, 97, E1051–E1059. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Meng, T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene 2019, 683, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, Q.; Xu, J.; Zhang, X.; Zhang, H.; Xiang, Y.; Fang, C.; Wang, T.; Xia, S.; Zhang, Q. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signaling. Sci. Rep. 2016, 6, 19910. [Google Scholar] [CrossRef]
- Zhu, X.-m.; Han, T.; Sargent, I.L.; Yin, G.-w.; Yao, Y.-q. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs. normal pregnancies. Am. J. Obstet. Gynecol. 2009, 200, e1-661.e7. [Google Scholar] [CrossRef]
- Zou, A.; Chen, B.; Li, Q.; Liang, Y. MiR-134 inhibits infiltration of trophoblast cells in placenta of patients with preeclampsia by decreasing ITGB1 expression. Eur. Rev. Med. Pharm. Sci. 2018, 22, 2199–2206. [Google Scholar]
- Bai, Y.; Yang, W.; Yang, H.-x.; Liao, Q.; Ye, G.; Fu, G.; Ji, L.; Xu, P.; Wang, H.; Li, Y.-x. Downregulated miR-195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS ONE 2012, 7, e38875. [Google Scholar] [CrossRef] [Green Version]
- Brkić, J.; Dunk, C.; O’Brien, J.; Fu, G.; Nadeem, L.; Wang, Y.-l.; Rosman, D.; Salem, M.; Shynlova, O.; Yougbaré, I. MicroRNA-218-5p promotes endovascular trophoblast differentiation and spiral artery remodeling. Mol. Ther. 2018, 26, 2189–2205. [Google Scholar] [CrossRef] [Green Version]
- Gunel, T.; Kamali, N.; Hosseini, M.K.; Gumusoglu, E.; Benian, A.; Aydinli, K. Regulatory effect of miR-195 in the placental dysfunction of preeclampsia. J. Matern. Fetal Neonatal Med. 2020, 33, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Li, Y.; Xu, Y. Decreased placental miR-126 expression and vascular endothelial growth factor levels in patients with pre-eclampsia. J. Int. Med Res. 2014, 42, 1243–1251. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Wang, N.; Xue, M.; Long, W.; Cheng, C.; Mi, C.; Gao, Z. A potential regulatory network among WDR86-AS1, miR-10b-3p, and LITAF is possibly involved in preeclampsia pathogenesis. Cell. Signal. 2019, 55, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; She, K.; Li, H.; Yuan, X.; Han, X.; Wang, Y. MicroRNA-454 contributes to sustaining the proliferation and invasion of trophoblast cells through inhibiting Nodal/ALK7 signaling in pre-eclampsia. Chem. Biol. Interact. 2019, 298, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yan, J. MicroRNA-454 is involved in regulating trophoblast cell proliferation, apoptosis, and invasion in preeclampsia by modulating the expression of ephrin receptor B4. Biomed. Pharmacother. 2018, 107, 746–753. [Google Scholar] [CrossRef]
- Xiaobo, Z.; Qizhi, H.; Zhiping, W.; Tao, D. Down-regulated miR-149-5p contributes to preeclampsia via modulating endoglin expression. Pregnancy Hypertens. 2019, 15, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Jiang, Z.; Yu, X.; Zhang, Y.; Sun, M.; Wang, W.; Ge, Z.; De, W.; Sun, L. MiR-101 regulates apoptosis of trophoblast HTR-8/SVneo cells by targeting endoplasmic reticulum (ER) protein 44 during preeclampsia. J. Hum. Hypertens. 2014, 28, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; MacIntyre, D.A.; Binkhamis, R.; Cook, J.; Sykes, L.; Bennett, P.R.; Terzidou, V. Maternal plasma miRNAs as potential biomarkers for detecting risk of small-for-gestational-age births. EBioMedicine 2020, 62, 103145. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J. MicroRNA-206 predicts raised fetal growth retardation risk through the interaction with vascular endothelial growth factor in pregnancies. Medicine 2020, 99, e18897. [Google Scholar] [CrossRef] [PubMed]
- Morales-Roselló, J.; García-Giménez, J.L.; Priego, L.M.; González-Rodríguez, D.; Mena-Mollá, S.; Catalá, A.M.; Loscalzo, G.; Buongiorno, S.; Jakaite, V.; Marín, A.P. MicroRNA-148b-3p and MicroRNA-25-3p Are Overexpressed in Fetuses with Late-Onset Fetal Growth Restriction. Fetal Diagn. Ther. 2020, 47, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Mouillet, J.-F.; Chu, T.; Hubel, C.A.; Nelson, D.M.; Parks, W.; Sadovsky, Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta 2010, 31, 781–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagliaferri, S.; Cepparulo, P.; Vinciguerra, A.; Campanile, M.; Esposito, G.; Maruotti, G.; Zullo, F.; Annunziato, L.; Pignataro, G. The microRNA cluster including miRNA16, miRNA27 and miRNA103 represents an early peripheral biomarker of fetal growth restriction. Authorea Prepr. 2020. [Google Scholar] [CrossRef]
- Wang, G.; Yu, J.; Yang, Y.; Liu, X.; Zhao, X.; Guo, X.; Duan, T.; Lu, C.; Kang, J. Whole-transcriptome sequencing uncovers core regulatory modules and gene signatures of human fetal growth restriction. Clin. Transl. Med. 2020, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The prediction of gestational hypertension, preeclampsia and fetal growth restriction via the first trimester screening of plasma exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972. [Google Scholar] [CrossRef]
- Ali, A.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. LIN28-let-7 axis regulates genes in immortalized human trophoblast cells by targeting the ARID3B-complex. FASEB J. 2019, 33, 12348–12363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awamleh, Z.; Han, V.K. Identification of miR-210-5p in human placentae from pregnancies complicated by preeclampsia and intrauterine growth restriction, and its potential role in the pregnancy complications. Pregnancy Hypertens. 2020, 19, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J. Reduced Cystathionine g-Lyase and Increased miR-21 Expression Are Associated with Increased Vascular Resistance in Growth-Restricted Pregnancies. Am. J. Pathol. 2013, 182. [Google Scholar] [CrossRef] [Green Version]
- Hromadnikova, I.; Kotlabova, K.; Hympanova, L.; Krofta, L. Cardiovascular and cerebrovascular disease associated microRNAs are dysregulated in placental tissues affected with gestational hypertension, preeclampsia and intrauterine growth restriction. PLoS ONE 2015, 10, e0138383. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Shen, Z.; Xu, Q.; Huang, X.; Chen, Q.; Li, D. Increased levels of microRNA-424 are associated with the pathogenesis of fetal growth restriction. Placenta 2013, 34, 624–627. [Google Scholar] [CrossRef]
- Li, L.; Huang, X.; He, Z.; Xiong, Y.; Fang, Q. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J. Cell. Mol. Med. 2019, 23, 4422–4433. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Cheng, Y.K.Y.; Wu, L.; Chaemsaithong, P.; Leung, M.B.W.; Chim, S.S.C.; Sahota, D.S.; Li, W.; Poon, L.C.Y.; Wang, C.C. Whole genome miRNA profiling revealed miR-199a as potential placental pathogenesis of selective fetal growth restriction in monochorionic twin pregnancies. Placenta 2020, 92, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Östling, H.; Kruse, R.; Helenius, G.; Lodefalk, M. Placental expression of microRNAs in infants born small for gestational age. Placenta 2019, 81, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Chakraborty, S.; Bhattacharya, A.; Biswas, A.; Ain, R. MicroRNA regulation of Transthyretin in trophoblast differentiation and Intra-Uterine Growth Restriction. Sci. Rep. 2017, 7, 16548. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Wu, W.; Xu, X.; Huang, L.; Gao, Q.; Chen, H.; Sun, H.; Xia, Y.; Sha, J.; Wang, X. miR-141 contributes to fetal growth restriction by regulating PLAG1 expression. PLoS ONE 2013, 8, e58737. [Google Scholar]
- Thamotharan, S.; Chu, A.; Kempf, K.; Janzen, C.; Grogan, T.; Elashoff, D.A.; Devaskar, S.U. Differential microRNA expression in human placentas of term intra-uterine growth restriction that regulates target genes mediating angiogenesis and amino acid transport. PLoS ONE 2017, 12, e0176493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Na, Q.; Song, W.-W.; Song, G.-Y. Altered expression of miR-518b and miR-519a in the placenta is associated with low fetal birth weight. Am. J. Perinatol. 2014, 31, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Chen, L.; He, J.; Lin, J. MicroRNA expression profiles and networks in placentas complicated with selective intrauterine growth restriction. Mol. Med. Rep. 2017, 16, 6650–6673. [Google Scholar] [CrossRef] [Green Version]
- Zou, Z.; He, Z.; Cai, J.; Huang, L.; Zhu, H.; Luo, Y. Potential role of microRNA-424 in regulating ERRγ to suppress trophoblast proliferation and invasion in fetal growth restriction. Placenta 2019, 83, 57–62. [Google Scholar] [CrossRef]
- Higashijima, A.; Miura, K.; Mishima, H.; Kinoshita, A.; Jo, O.; Abe, S.; Hasegawa, Y.; Miura, S.; Yamasaki, K.; Yoshida, A. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat. Diagn. 2013, 33, 214–222. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34 (Suppl. 2), W451–W454. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Chang, G.; Mouillet, J.F.; Mishima, T.; Chu, T.; Sadovsky, E.; Coyne, C.B.; Parks, W.T.; Surti, U.; Sadovsky, Y. Expression and trafficking of placental microRNAs at the feto-maternal interface. FASEB J. 2017, 31, 2760–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floris, I.; Kraft, J.D.; Altosaar, I. Roles of microRNA across prenatal and postnatal periods. Int. J. Mol. Sci. 2016, 17, 1994. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J. Fetal origins of coronary heart disease. BMJ 1995, 311, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.-P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jens, M.; Rajewsky, N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015, 16, 113–126. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, W.; Yuan, Y.; Bai, Y.; Sun, Y.; Zhu, W.; Du, Z. MicroRNAs tend to synergistically control expression of genes encoding extensively-expressed proteins in humans. PeerJ 2017, 5, e3682. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Finley, S.D. Mechanistic insight into activation of MAPK signaling by pro-angiogenic factors. BMC Syst. Biol. 2018, 12, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravikumar, G.; Mukhopadhyay, A.; Mani, C.; Kocchar, P.; Crasta, J.; Thomas, T.; Dwarkanath, P.; Thomas, A.; Kurpad, A.V.; Sridhar, T.S. Placental expression of angiogenesis-related genes and their receptors in IUGR pregnancies: Correlation with fetoplacental and maternal parameters. J. Matern. Fetal Neonatal Med. 2019, 33, 3954–3961. [Google Scholar] [CrossRef]
- Wang, Y. Vascular Biology of the Placenta. In Colloquium Series on Integrated Systems Physiology: From Molecule to Function, 2010; Morgan & Claypool Life Sciences: Copenhagen, Denmark, 2010; pp. 1–98. [Google Scholar]
- White, V.; Jawerbaum, A.; Mazzucco, M.B.; Gauster, M.; Desoye, G.; Hiden, U. IGF2 stimulates fetal growth in a sex-and organ-dependent manner. Pediatr. Res. 2018, 83, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Mu, Q.; Huang, H. The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int. 2018, 2018, 4217259. [Google Scholar] [CrossRef] [Green Version]
- Amirabad, A.D.; Ramasamy, P.; Wierz, M.; Nordström, K.; Kessler, S.M.; Schulz, M.H.; Simon, M. Transgenic expression of the RNA binding protein IMP2 stabilizes miRNA targets in murine microsteatosis. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2018, 1864, 3099–3108. [Google Scholar] [CrossRef]
- Degrauwe, N.; Suvà, M.-L.; Janiszewska, M.; Riggi, N.; Stamenkovic, I. IMPs: An RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016, 30, 2459–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, X.; Zhang, L.; Shi, Y.; Wang, J.; Yan, H. Wnt/β-catenin signaling pathway in trophoblasts and abnormal activation in preeclampsia. Mol. Med. Rep. 2017, 16, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Devi, H.L.; Kumar, S.; Konyak, Y.; Bharati, J.; Bhimte, A.; Pandey, Y.; Kumar, K.; Paul, A.; Kala, A.; Samad, H. Expression and functional role of fibroblast growth factors (FGF) in placenta during different stages of pregnancy in water buffalo (Bubalus bubalis). Theriogenology 2020, 143, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Pearce, W.J. Multifunctional angiogenic factors: Add GnRH to the list. Focus on “Gonadotropin-releasing hormone-regulated chemokine expression in human placentation”. Am. J. Physiol. Cell Physiol. 2009, 297, C4–C5. [Google Scholar] [CrossRef] [Green Version]
- Rab, A.; Szentpéteri, I.; Kornya, L.; Börzsönyi, B.; Demendi, C.; Joó, J.G. Placental gene expression patterns of epidermal growth factor in intrauterine growth restriction. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 170, 96–99. [Google Scholar] [CrossRef]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Shin, N.; Chen, S.; Lei, J.; Burd, I.; Wang, X. Is there a definite relationship between placental mTOR signaling and fetal growth? Biol. Reprod. 2020, 103, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.; Choi, Y.; Shin, Y.; Ahn, G.; Kim, K.H.; Kim, W.; Lee, H.; Kim, S. Expression of TGF-ß signaling proteins in normal placenta and gestational trophoblastic disease. Histol. Histopathol. 2007, 22, 227–234. [Google Scholar]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varelas, X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 2014, 141, 1614–1626. [Google Scholar] [CrossRef] [Green Version]
- Soncin, F.; Parast, M.M. Role of Hippo signaling pathway in early placental development. Proc. Natl. Acad. Sci. USA 2020, 117, 20354–20356. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Z.; Zeng, X.; Wang, J.; Zhang, L.; Song, W.; Shi, Y. Wnt/β-catenin signaling pathway in severe preeclampsia. J. Mol. Histol. 2018, 49, 317–327. [Google Scholar] [CrossRef]
- Bavelloni, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A current overview. Anticancer Res. 2017, 37, 6511–6521. [Google Scholar]
- Soares, M.J.; Iqbal, K.; Kozai, K. Hypoxia and placental development. Birth Defects Res. 2017, 109, 1309–1329. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Liu, J.; Yang, Q.; Liu, Y.; Jia, W.; Zhang, X.; Li, Y.-X.; Shao, X.; Wang, Y.-L. MicroRNA-210 regulates placental adaptation to maternal hypoxic stress during pregnancy. Biol. Reprod. 2020, 104, 418–429. [Google Scholar] [CrossRef]








| Name | Regulation | Source | References |
|---|---|---|---|
| let-7a, let-7c, miR-103, miR-122, miR-1233, miR-125a, miR-125b, miR-130b, miR-1323, miR-133a, miR-136, miR-141, miR-143, miR-152, miR-155, miR-181a, miR-182, miR-192, miR-193b, miR-21, miR-210, miR-215, miR-221, miR-24, miR-26a, miR-29a, miR-342, miR-423, miR-494, miR-495, miR-512, miR-515, miR-516a, miR-516b, miR-517b, miR-517c, miR-518b, miR-518e, miR-518f, miR-519a, miR-519d, miR-520a, miR-520c, miR-520d, miR-520g, miR-520h, miR-521, miR-525, miR-526b, miR-542, miR-574, miR-628, miR-629, miR-650, miR-758, miR-92a | Upregulated | Maternal circulation | [41,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67] |
| miR-185, let-7d, miR-126, miR-1260, miR-1272, miR-144, miR-196b, miR-19b1, miR-223, miR-320c, miR-92a1, miR-766, miR-573, miR-409, miR-18a | Downregulated | Maternal circulation | [41,49,51,60,61,63,66,68,69] |
| let-7c, let-7d, miR-122, miR-125b, miR-134, miR-136, miR-141, miR-148a, miR-151, miR-152, miR-155, miR-16, miR-17, miR-181a, miR-193b-star, miR-20a, miR-20b, miR-21, miR-210, miR-221, miR-222, miR-25, miR-26b, miR-27a-star, miR-296, miR-29b, miR-30a-star, miR-31, miR-320a, miR-335, miR-362, miR-365a, miR-423, miR-431, miR-4421, miR-516b, miR-517-star, miR-518a, miR-518b, miR-519e, miR-520a, miR-524, miR-584, miR-638 | Upregulated | Placenta | [10,45,62,66,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89] |
| miR-542, miR-584, miR-17, miR-18a, miR-223, miR-19b1, miR-144, miR-126, miR-92a1, miR-1247, miR-204, miR-590, miR-1, miR-363, miR-150, miR-218, miR-32, miR-328, miR-625, miR-19a, miR-18b, miR-154, miR-411, miR-101, miR-195, miR-135b, miR-454, miR-374, miR-379, miR-149, miR-377, miR-10b, miR-450, miR-34c, miR-500, miR-139 | Downregulated | Placenta | [66,73,87,88,90,91,92,93,94,95,96,97,98] |
| Name | Regulation | Where | References |
|---|---|---|---|
| let-7d, miR-103, miR-1306, miR-141, miR-148b, miR-16, miR-191, miR-200c, miR-205, miR-206, miR-224, miR-25, miR-27b, miR-27a-star, miR-30d, miR-335, miR-374a, miR-424, miR-432, miR-451, miR-491, miR-517a, miR-518b, miR-518e, miR-524, miR-93 | Upregulated | Maternal circulation | [99,100,101,102,103,104] |
| miR-4454, miR-520a, miR-7975 | Downregulated | Maternal circulation | [99,105] |
| let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-10b, miR-124, miR-141, miR-193b, miR-193b-star, miR-199a, miR-21, miR-210, miR-338, miR-34b, miR-363, miR-365a, miR-3679, miR-373, miR-424, miR-4287, miR-499a, miR-519a, miR-523, miR-572, miR-574, miR-590, miR-623, miR-664b, miR-758 | Upregulated | Placenta | [10,106,107,108,109,110,111,112,113,114,115,116,117,118,119] |
| miR-526b, miR-5581, miR-519d, miR-519e, miR-520h, miR-515, miR-516b, miR-5189, miR-518b, miR-4535, miR-4743, miR-379-star, miR-380, miR-3622b, miR-370, miR-1323, miR-1, miR-105 | Downregulated | Placenta | [113,118,120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, A.; Hadlich, F.; Abbas, M.W.; Iqbal, M.A.; Tesfaye, D.; Bouma, G.J.; Winger, Q.A.; Ponsuksili, S. MicroRNA–mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers. Int. J. Mol. Sci. 2021, 22, 2313. https://doi.org/10.3390/ijms22052313
Ali A, Hadlich F, Abbas MW, Iqbal MA, Tesfaye D, Bouma GJ, Winger QA, Ponsuksili S. MicroRNA–mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers. International Journal of Molecular Sciences. 2021; 22(5):2313. https://doi.org/10.3390/ijms22052313
Chicago/Turabian StyleAli, Asghar, Frieder Hadlich, Muhammad W. Abbas, Muhammad A. Iqbal, Dawit Tesfaye, Gerrit J. Bouma, Quinton A. Winger, and Siriluck Ponsuksili. 2021. "MicroRNA–mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers" International Journal of Molecular Sciences 22, no. 5: 2313. https://doi.org/10.3390/ijms22052313