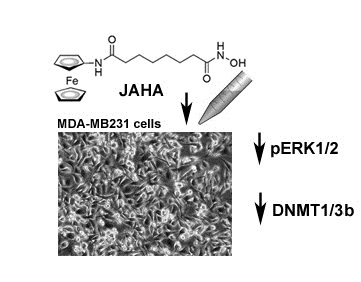
The Histone Deacetylase Inhibitor JAHA Down-Regulates pERK and Global DNA Methylation in MDA-MB231 Breast Cancer Cells
Abstract
:1. Introduction

2. Results and Discussion



3. Experimental Section
3.1. Cell Culture and JAHA Treatment
3.2. Western Blot
3.3. Methylation-Sensitive Arbitrarily-Primed (MeSAP)-PCR
3.4. ELISA Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Spencer, J.; Amin, J.; Wang, M.; Packham, G.; Alwi, S.S.; Tizzard, G.J.; Coles, S.J.; Paranal, R.M.; Bradner, J.E.; Heightman, T.D. Synthesis and biological evaluation of JAHAs: Ferrocene-based histone deacetylase inhibitors. ACS Med. Chem. Lett. 2011, 2, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.; Morgan, M.P.; Marmion, C.J. A novel anti-cancer bifunctional platinum drug candidate with dual DNA binding and histone deacetylase inhibitory activity. Chem. Commun. 2009, 44, 6735–6737. [Google Scholar] [CrossRef] [PubMed]
- Can, D.; Peindy N’Dongo, H.W.; Spingler, B.; Schmutz, P.; Raposinho, P.; Santos, I.; Alberto, R. The [(Cp)M(CO)3] (M = Re, 99 mTc) building block for imaging agents and bioinorganic probes: Perspectives and limitations. Chem. Biodivers. 2012, 9, 1849–1866. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.; Amin, J.; Boddiboyena, R.; Packham, G.; Cavell, B.E.; Syed Alwi, S.S.; Paranal, R.M.; Heightman, T.D.; Wang, M.; Marsden, B.; et al. Click JAHAs: Conformationally restricted ferrocene-based histone deacetylase inhibitors. MedChemComm 2012, 3, 61–64. [Google Scholar] [CrossRef]
- Ye, R.R.; Ke, Z.F.; Tan, C.P.; He, L.; Ji, L.N.; Mao, Z.W. Histone-deacetylase-targeted fluorescent ruthenium(II) polypyridyl complexes as potent anticancer agents. Chemistry 2013, 19, 10160–10169. [Google Scholar] [CrossRef] [PubMed]
- De Jesús Cázares-Marinero, J.; Top, S.; Vessières, A.; Jaouen, G. Synthesis and antiproliferative activity of hydroxyferrocifen hybrids against triple-negative breast cancer cells. Dalton Trans. 2014, 43, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.R.; Tan, C.P.; Lin, Y.N.; Ji, L.N.; Mao, Z.W. A phosphorescent rhenium(I) histone deacetylase inhibitor: Mitochondrial targeting and paraptosis induction. Chem. Commun. 2015, 51, 8353–8356. [Google Scholar] [CrossRef] [PubMed]
- Librizzi, M.; Longo, A.; Chiarelli, R.; Amin, J.; Spencer, J.; Luparello, C. Cytotoxic effects of Jay Amin hydroxamic acid (JAHA), a ferrocene-based class I histone deacetylase inhibitor, on triple-negative MDA-MB231 breast cancer cells. Chem. Res. Toxicol. 2012, 25, 2608–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellarosa, D.; Bressan, A.; Bigioni, M.; Parlani, M.; Maggi, C.A.; Binaschi, M. SAHA/Vorinostat induces the expression of the CD137 receptor/ligand system and enhances apoptosis mediated by soluble CD137 receptor in a human breast cancer cell line. Int. J. Oncol. 2012, 41, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5, A006098. [Google Scholar] [CrossRef] [PubMed]
- Bali, P.; Pranpat, M.; Swaby, R.; Fiskus, W.; Yamaguchi, H.; Balasis, M.; Rocha, K.; Wang, H.G.; Richon, V.; Bhalla, K. Activity of suberoylanilide hydroxamic acid against human breast cancer cells with amplification of her-2. Clin. Cancer Res. 2005, 11, 6382–6389. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, D.; Wu, H.; Liu, H.; Zhou, H.; Gu, R.; Zhang, R.; Zhang, S.; Wu, G. Anticancer activity of SAHA, a potent histone deacetylase inhibitor, in NCI-H460 human large-cell lung carcinoma cells in vitro and in vivo. Int. J. Oncol. 2014, 44, 451–458. [Google Scholar] [PubMed]
- Yang, B.; Yu, D.; Liu, J.; Yang, K.; Wu, G.; Liu, H. Antitumor activity of SAHA, a novel histone deacetylase inhibitor, against murine B cell lymphoma A20 cells in vitro and in vivo. Tumour Biol. 2015, 36, 5051–5061. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.C.; Cho, C.Y.; Hung, W.C. Silencing of the metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b-induced promoter methylation. Cancer Res. 2006, 66, 8413–8420. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gorelik, G.J.; Strickland, F.M.; Richardson, B.C. Decreased ERK and JNK signaling contribute to gene overexpression in “senescent” CD4+CD28- T cells through epi-genetic mechanisms. J. Leukoc. Biol. 2010, 87, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Abujamra, A.L.; Loew, J.E.; Forman, L.W.; Perrine, S.P.; Faller, D.V. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res. 2011, 31, 2723–2732. [Google Scholar] [PubMed]
- Caradonna, F.; Barbata, G.; Sciandrello, G. Genomewide hypomethylation and PTHrP gene hypermethylation as a model for the prediction of cancer risk in rheumatoid arthritis. In Novel Aspects of PTHrP Physiopathology; Luparello, C., Ed.; Nova Science Publisher: Happauge, NY, USA, 2007; pp. 305–319. [Google Scholar]
- Naselli, F.; Catanzaro, I.; Bellavia, D.; Perez, A.; Sposito, L.; Caradonna, F. Role and importance of polymorphisms with respect to DNA methylation for the expression of CYP2E1 enzyme. Gene 2014, 536, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Fuks, F.; Burgers, W.A.; Brehm, A.; Hughes-Davies, L.; Kouzarides, T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000, 24, 88–91. [Google Scholar] [PubMed]
- Arzenani, M.K.; Zade, A.E.; Ming, Y.; Vijverberg, S.J.; Zhang, Z.; Khan, Z.; Sadique, S.; Kallenbach, L.; Hu, L.; Vukojević, V.; et al. Genomic DNA hypomethylation by histone deacetylase inhibition implicates DNMT1 nuclear dynamics. Mol. Cell Biol. 2011, 31, 4119–4128. [Google Scholar] [CrossRef] [PubMed]
- Chiarelli, R.; Agnello, M.; Roccheri, M.C. Sea urchin embryos as a model system for studying autophagy induced by cadmium stress. Autophagy 2011, 7, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Uehara, N.; Kanematsu, S.; Miki, H.; Yoshizawa, K.; Tsubura, A. Requirement of p38 MAPK for a cell-death pathway triggered by vorinostat in MDA-MB-231 human breast cancer cells. Cancer Lett. 2012, 315, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, F.; Ballestar, E.; Ropero, S.; Esteller, M. Discovery of epigenetically silenced genes by methylated DNA immunoprecipitation in colon cancer cells. Cancer Res. 2007, 67, 11481–11486. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA methyltransferases: A novel target for prevention and therapy. Front. Oncol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Librizzi, M.; Chiarelli, R.; Bosco, L.; Sansook, S.; Gascon, J.M.; Spencer, J.; Caradonna, F.; Luparello, C. The Histone Deacetylase Inhibitor JAHA Down-Regulates pERK and Global DNA Methylation in MDA-MB231 Breast Cancer Cells. Materials 2015, 8, 7041-7047. https://doi.org/10.3390/ma8105358
Librizzi M, Chiarelli R, Bosco L, Sansook S, Gascon JM, Spencer J, Caradonna F, Luparello C. The Histone Deacetylase Inhibitor JAHA Down-Regulates pERK and Global DNA Methylation in MDA-MB231 Breast Cancer Cells. Materials. 2015; 8(10):7041-7047. https://doi.org/10.3390/ma8105358
Chicago/Turabian StyleLibrizzi, Mariangela, Roberto Chiarelli, Liana Bosco, Supojjanee Sansook, Jose M. Gascon, John Spencer, Fabio Caradonna, and Claudio Luparello. 2015. "The Histone Deacetylase Inhibitor JAHA Down-Regulates pERK and Global DNA Methylation in MDA-MB231 Breast Cancer Cells" Materials 8, no. 10: 7041-7047. https://doi.org/10.3390/ma8105358