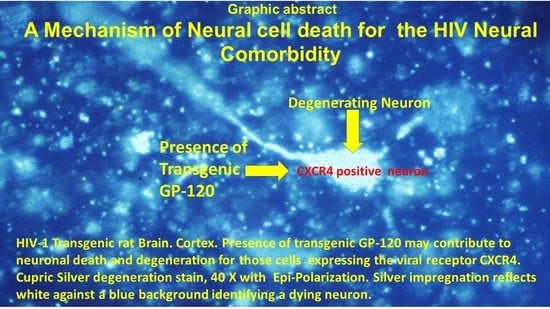
The HIV-1 Transgenic Rat: Relevance for HIV Noninfectious Comorbidity Research
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Processing
2.3. Immunocytochemistry for Transgene Products gp120 and Tat
2.4. Immunochemistry for CXCR4
2.5. Special Stain
2.6. Routine Stain
2.7. Slide Review of H&E Serial Sections
2.8. Slide Review Silver Stain
2.9. Slide Review of CXCR4 Immunostaining
2.10. Imaging
3. Results
3.1. Cortical Pathology
3.2. Hippocampal Pathology
3.3. Striatal Pathology
3.4. Immunohistochemical Identification of gp120 and Tat in the Brain
3.5. Spleen Pathology
3.5.1. Immunohistochemical Identification of gp120 and Tat
3.5.2. Identification of CXCR4 in the Spleen
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 15035. [Google Scholar] [CrossRef]
- Ghosn, J.; Taiwo, B.; Seedat, S.; Autran, B.; Katlama, C. HIV. Lancet 2018, 392, 685–697. [Google Scholar] [CrossRef]
- Heron, J.E.; Norman, S.M.; Yoo, J.; Lembke, K.; O’Connor, C.C.; Weston, C.E.; Gracey, D.M. The prevalence and risk of non-infectious comorbidities in HIV-infected and non-HIV infected men attending general practice in Australia. PLoS ONE 2019, 14, e0223224. [Google Scholar] [CrossRef] [PubMed]
- Hjalte, F.; Calara, P.S.; Blaxhult, A.; Helleberg, M.; Wallace, K.; Lindgren, P. Excess costs of non-infectious comorbidities among people living with HIV—estimates from Denmark and Sweden. AIDS Care 2018, 30, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Hernández, I.; Barzallo, J.; Beltrán, S.; Castillo, A.; Cevallos, N.; Hernández, P.; López, C.; Vera, R.; Yerovi, G.; Mendoza, A.; et al. Increased incidences of noninfectious comorbidities among aging populations living with human immunodeficiency virus in Ecuador: A multicenter retrospective analysis. HIV/AIDS (Auckland N.Z.) 2019, 11, 55–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Kindi, S.G.; ElAmm, C.; Ginwalla, M.; Mehanna, E.; Zacharias, M.; Benatti, R.; Oliveira, G.H.; Longenecker, C.T. Heart failure in patients with human immunodeficiency virus infection: Epidemiology and management disparities. Int. J. Cardiol. 2016, 218, 43–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbein, G.; Coaquette, A.; Perez-Bercoff, D.; Pancino, G. Macrophage activation and HIV infection: Can the Trojan horse turn into a fortress? Curr. Mol. Med. 2002, 2, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Fantuzzi, L.; Conti, L.; Gauzzi, M.C.; Eid, P.; Del Cornò, M.; Varano, B.; Canini, I.; Belardelli, F.; Gessani, S. Regulation of chemokine/cytokine network during in vitro differentiation and HIV-1 infection of human monocytes: Possible importance in the pathogenesis of AIDS. J. Leukoc. Biol. 2000, 68, 391–399. [Google Scholar]
- Fantuzzi, L.; Belardelli, F.; Gessani, S. Monocyte/macrophage-derived CC chemokines and their modulation by HIV-1 and cytokines: A complex network of interactions influencing viral replication and AIDS pathogenesis. J. Leukoc. Biol. 2003, 74, 719–725. [Google Scholar] [CrossRef]
- Herbein, G.; Keshav, S.; Collin, M.; Montaner, L.J.; Gordon, S. HIV-1 induces tumour necrosis factor and IL-1 gene expression in primary human macrophages independent of productive infection. Clin. Exp. Immunol. 1994, 95, 442–449. [Google Scholar] [CrossRef]
- Wahl, S.M.; Greenwell-Wild, T.; Peng, G.; Ma, G.; Orenstein, J.M.; Vazquez, N. Viral and host cofactors facilitate HIV-1 replication in macrophages. J. Leukoc. Biol. 2003, 74, 726–735. [Google Scholar] [CrossRef]
- Merrill, J.E.; Koyanagi, Y.; Chen, I.S. Interleukin-1 and tumor necrosis factor alpha can be induced from mononuclear phagocytes by human immunodeficiency virus type 1 binding to the CD4 receptor. J. Virol. 1989, 63, 4404–4408. [Google Scholar] [CrossRef] [Green Version]
- Merrill, J.E.; Koyanagi, Y.; Zack, J.; Thomas, L.; Martin, F.; Chen, I.S. Induction of interleukin-1 and tumor necrosis factor alpha in brain cultures by human immunodeficiency virus type 1. J. Virol. 1992, 66, 2217–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lathey, J.L.; Kanangat, S.; Rouse, B.T.; Agosti, J.M.; Spector, S.A. Dysregulation of cytokine expression in monocytes from HIV-positive individuals. J. Leukoc. Biol. 1994, 56, 347–352. [Google Scholar] [CrossRef]
- Bergamini, A.; Faggioli, E.; Bolacchi, F.; Gessani, S.; Cappannoli, L.; Uccella, I.; Demin, F.; Capozzi, M.; Cicconi, R.; Placido, R.; et al. Enhanced Production of Tumor Necrosis Factor-α and Interleukin-6 Due to Prolonged Response to Lipopolysaccharide in Human Macrophages Infected In Vitro with Human Immunodeficiency Virus Type 1. J. Infect. Dis. 1999, 179, 832–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emilie, D.; Peuchmaur, M.; Maillot, M.C.; Crevon, M.C.; Brousse, N.; Delfraissy, J.F.; Dormont, J.; Galanaud, P. Production of interleukins in human immunodeficiency virus-1-replicating lymph nodes. J. Clin. Investig. 1990, 86, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Agostini, C.; Trentin, L.; Zambello, R.; Bulian, P.; Caenazzo, C.; Cipriani, A.; Cadrobbi, P.; Garbisa, S.; Semenzato, G. Release of granulocyte-macrophage colony-stimulating factor by alveolar macrophages in the lung of HIV-1-infected patients. A mechanism accounting for macrophage and neutrophil accumulation. J. Immunol. 1992, 149, 3379–3385. [Google Scholar] [PubMed]
- Chehimi, J.; Starr, S.E.; Frank, I.; D′Andrea, A.; Ma, X.; MacGregor, R.R.; Sennelier, J.; Trinchieri, G. Impaired interleukin 12 production in human immunodeficiency virus-infected patients. J. Exp. Med. 1994, 179, 1361–1366. [Google Scholar] [CrossRef]
- Yoo, J.; Chen, H.; Kraus, T.; Hirsch, D.; Polyak, S.; George, I.; Sperber, K. Altered cytokine production and accessory cell function after HIV-1 infection. J. Immunol. 1996, 157, 1313–1320. [Google Scholar]
- Chougnet, C.; Thomas, E.; Landay, A.L.; Kessler, H.A.; Buchbinder, S.; Scheer, S.; Shearer, G.M. CD40 ligand and IFN-gamma synergistically restore IL-12 production in HIV-infected patients. Eur. J. Immunol. 1998, 28, 646–656. [Google Scholar] [CrossRef]
- Chougnet, C.; Wynn, T.A.; Clerici, M.; Landay, A.L.; Kessler, H.A.; Rusnak, J.; Melcher, G.P.; Sher, A.; Shearer, G.M. Molecular analysis of decreased interleukin-12 production in persons infected with human immunodeficiency virus. J. Infect. Dis. 1996, 174, 46–53. [Google Scholar] [CrossRef]
- Akridge, R.E.; Oyafuso, L.K.; Reed, S.G. IL-10 is induced during HIV-1 infection and is capable of decreasing viral replication in human macrophages. J. Immunol. 1994, 153, 5782–5789. [Google Scholar]
- Bergamini, A.; Bolacchi, F.; Faggioli, E.; Placido, R.; Vendetti, S.; Cappannoli, L.; Ventura, L.; Cerasari, G.; Uccella, I.; Andreoni, M.; et al. HIV-1 does not alter in vitro and in vivo IL-10 production by human monocytes and macrophages. Clin. Exp. Immunol. 1998, 112, 105–111. [Google Scholar] [CrossRef]
- Dereuddre-Bosquet, N.; Clayette, P.; Martin, M.; Benveniste, O.; Fretier, P.; Jaccard, P.; Vaslin, B.; Lebeaut, A.; Dormont, D. Lack of interleukin 10 expression in monocyte-derived macrophages in response to in vitro infection by HIV type 1 isolates. AIDS Res. Human Retrovir. 1997, 13, 961–966. [Google Scholar] [CrossRef]
- Fantuzzi, G. Lessons from interleukin-deficient mice: The interleukin-1 system. Acta Physiol. Scand. 2001, 173, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.L.; Meltzer, M.S. Induction of IFN-alpha by HIV-1 in monocyte-enriched PBMC requires gp120-CD4 interaction but not virus replication. J. Immunol. 1993, 151, 2208–2216. [Google Scholar]
- Szebeni, J.; Dieffenbach, C.; Wahl, S.M.; Venkateshan, C.N.; Yeh, A.; Popovic, M.; Gartner, S.; Wahl, L.M.; Peterfy, M.; Friedman, R.M. Induction of alpha interferon by human immunodeficiency virus type 1 in human monocyte-macrophage cultures. J. Virol. 1991, 65, 6362–6364. [Google Scholar] [CrossRef] [Green Version]
- Gessani, S.; Puddu, P.; Varano, B.; Borghi, P.; Conti, L.; Fantuzzi, L.; Belardelli, F. Induction of beta interferon by human immunodeficiency virus type 1 and its gp120 protein in human monocytes-macrophages: Role of beta interferon in restriction of virus replication. J. Virol. 1994, 68, 1983–1986. [Google Scholar] [CrossRef] [Green Version]
- Gruber, M.F.; Weih, K.A.; Boone, E.J.; Smith, P.D.; Clouse, K.A. Endogenous macrophage CSF production is associated with viral replication in HIV-1-infected human monocyte-derived macrophages. J. Immunol. 1995, 154, 5528–5535. [Google Scholar]
- Schmidtmayerova, H.; Nottet, H.S.; Nuovo, G.; Raabe, T.; Flanagan, C.R.; Dubrovsky, L.; Gendelman, H.E.; Cerami, A.; Bukrinsky, M.; Sherry, B. Human immunodeficiency virus type 1 infection alters chemokine beta peptide expression in human monocytes: Implications for recruitment of leukocytes into brain and lymph nodes. Proc. Natl. Acad. Sci. USA 1996, 93, 700–704. [Google Scholar] [CrossRef] [Green Version]
- Canque, B.; Rosenzwajg, M.; Gey, A.; Tartour, E.; Fridman, W.H.; Gluckman, J.C. Macrophage inflammatory protein-1alpha is induced by human immunodeficiency virus infection of monocyte-derived macrophages. Blood 1996, 87, 2011–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denis, M.; Ghadirian, E. Alveolar macrophages from subjects infected with HIV-1 express macrophage inflammatory protein-1 alpha (MIP-1 alpha): Contribution to the CD8+ alveolitis. Clin. Exp. Immunol. 1994, 96, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Choe, W.; Volsky, D.J.; Potash, M.J. Induction of rapid and extensive beta-chemokine synthesis in macrophages by human immunodeficiency virus type 1 and gp120, independently of their coreceptor phenotype. J. Virol. 2001, 75, 10738–10745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, M.E.; Jaworowski, A.; Hearps, A.C. The HIV Reservoir in Monocytes and Macrophages. Front. Immunol. 2019, 10, 1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruize, Z.; Kootstra, N.A. The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Front. Immunol. 2019, 10, 2828. [Google Scholar] [CrossRef] [Green Version]
- Falcinelli, S.D.; Ceriani, C.; Margolis, D.M.; Archin, N.M. New Frontiers in Measuring and Characterizing the HIV Reservoir. Front. Immunol. 2019, 10, 2878. [Google Scholar] [CrossRef] [Green Version]
- Zicari, S.; Sessa, L.; Cotugno, N.; Ruggiero, A.; Morrocchi, E.; Concato, C.; Rocca, S.; Zangari, P.; Manno, E.C.; Palma, P. Immune Activation, Inflammation, and Non-AIDS Co-Morbidities in HIV-Infected Patients under Long-Term ART. Viruses 2019, 11, 200. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.; Kossenkov, A.V.; Knecht, V.R.; Showe, L.C.; Ratcliffe, S.J.; Montaner, L.J.; Tebas, P.; Collman, R.G. Evidence for Persistent Monocyte and Immune Dysregulation After Prolonged Viral Suppression Despite Normalization of Monocyte Subsets, sCD14 and sCD163 in HIV-Infected Individuals. Pathog. Immun. 2019, 4, 324–362. [Google Scholar] [CrossRef]
- Hong, S.; Banks, W. Role of the Immune System in HIV-associated Neuroinflammation and Neurocognitive Implications. Brain Behav. Immun. 2014, 45, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Davinelli, S.; Scapagnini, G.; Denaro, F.; Calabrese, V.; Benedetti, F.; Krishnan, S.; Curreli, S.; Bryant, J.; Zella, D. Altered expression pattern of Nrf2/HO-1 axis during accelerated-senescence in HIV-1 transgenic rat. Biogerontology 2014, 15, 449–461. [Google Scholar] [CrossRef]
- Klotman, P.E.; Rappaport, J.; Ray, P.; Kopp, J.B.; Franks, R.; Bruggeman, L.A.; Notkins, A.L. Transgenic models of HIV-1. Aids 1995, 9, 313–324. [Google Scholar] [CrossRef]
- Hanna, Z.; Kay, D.G.; Cool, M.; Jothy, S.; Rebai, N.; Jolicoeur, P. Transgenic mice expressing human immunodeficiency virus type 1 in immune cells develop a severe AIDS-like disease. J. Virol. 1998, 72, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Reid, W.; Sadowska, M.; Denaro, F.; Rao, S.; Foulke, J.; Hayes, N.; Jones, O.; Doodnauth, D.; Davis, H.; Sill, A.; et al. An HIV-1 transgenic rat that develops HIV-related pathology and immunologic dysfunction. Proc. Natl. Acad. Sci. USA 2001, 98, 9271–9276. [Google Scholar] [CrossRef] [Green Version]
- Yedavalli, V.S.; Benkirane, M.; Jeang, K.T. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 2003, 278, 6404–6410. [Google Scholar] [CrossRef] [Green Version]
- Reid, W.C.; Ibrahim, W.G.; Kim, S.J.; Denaro, F.; Casas, R.; Lee, D.E.; Maric, D.; Hammoud, D.A. Characterization of neuropathology in the HIV-1 transgenic rat at different ages. J. Neuroimmunol. 2016, 292, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Mazzucchelli, R.; Amadio, M.; Curreli, S.; Denaro, F.; Bemis, K.; Reid, W.; Bryant, J.; Riva, A.; Galli, M.; Zella, D. Establishment of an ex vivo model of monocytes-derived macrophages differentiated from peripheral blood mononuclear cells (PBMCs) from HIV-1 transgenic rats. Mol. Immunol. 2004, 41, 979–984. [Google Scholar] [CrossRef]
- Keppler, O.T.; Yonemoto, W.; Welte, F.J.; Patton, K.S.; Iacovides, D.; Atchison, R.E.; Ngo, T.; Hirschberg, D.L.; Speck, R.F.; Goldsmith, M.A. Susceptibility of Rat-Derived Cells to Replication by Human Immunodeficiency Virus Type 1. J. Virol. 2001, 75, 8063. [Google Scholar] [CrossRef] [Green Version]
- Pleskoff, O.; Sol, N.; Labrosse, B.; Alizon, M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR-4 (fusin). J. Virol. 1997, 71, 3259–3262. [Google Scholar] [CrossRef] [Green Version]
- Repunte-Canonigo, V.; Lefebvre, C.; George, O.; Kawamura, T.; Morales, M.; Koob, G.F.; Califano, A.; Masliah, E.; Sanna, P.P. Gene expression changes consistent with neuroAIDS and impaired working memory in HIV-1 transgenic rats. Mol. Neurodegener. 2014, 9, 26. [Google Scholar] [CrossRef] [Green Version]
- Nesil, T.; Cao, J.; Yang, Z.; Chang, S.L.; Li, M.D. Nicotine attenuates the effect of HIV-1 proteins on the neural circuits of working and contextual memories. Mol. Brain 2015, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Maric, D.; Denaro, F.; Ibrahim, W.; Mason, R.; Kumar, A.; Hammoud, D.A.; Reid, W. Nitrosative Stress Is Associated with Dopaminergic Dysfunction in the HIV-1 Transgenic Rat. Am. J. Pathol. 2019, 189, 1375–1385. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 7th ed.; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Switzer, R.C. Application of Silver Degeneration Stains for Neurotoxicity Testing. Toxicol. Pathol. 2000, 28, 70–83. [Google Scholar] [CrossRef]
- Tenkova, T.I.; Goldberg, M.P. A Modified Silver Technique (de Olmos Stain) for Assessment of Neuronal and Axonal Degeneration, in Neuroprotection Methods and Protocols; Borsello, T., Ed.; Humana Press: Totowa, NJ, USA, 2007; pp. 31–39. [Google Scholar]
- Trecki, J.; Brailoiu, G.C.; Unterwald, E.M. Localization of CXCR4 in the forebrain of the adult rat. Brain Res. 2010, 1315, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Mensah, P.; Deadwyler, S. The caudate nucleus of the rat: Cell types and the demonstration of a commissural system. J. Anat. 1974, 117, 281–293. [Google Scholar]
- Peng, J.; Vigorito, M.; Liu, X.; Zhou, D.; Wu, X.; Chang, S.L. The HIV-1 transgenic rat as a model for HIV-1 infected individuals on HAART. J. Neuroimmunol. 2010, 218, 94–101. [Google Scholar] [CrossRef]
- Secchiero, P.; Zella, D.; Capitani, S.; Gallo, R.C.; Zauli, G. Extracellular HIV-1 Tat Protein Up-Regulates the Expression of Surface CXC-Chemokine Receptor 4 in Resting CD4+ T Cells. J. Immunol. 1999, 162, 2427. [Google Scholar]















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denaro, F.; Benedetti, F.; Worthington, M.D.; Scapagnini, G.; Krauss, C.C.; Williams, S.; Bryant, J.; Davis, H.; Latinovic, O.S.; Zella, D. The HIV-1 Transgenic Rat: Relevance for HIV Noninfectious Comorbidity Research. Microorganisms 2020, 8, 1643. https://doi.org/10.3390/microorganisms8111643
Denaro F, Benedetti F, Worthington MD, Scapagnini G, Krauss CC, Williams S, Bryant J, Davis H, Latinovic OS, Zella D. The HIV-1 Transgenic Rat: Relevance for HIV Noninfectious Comorbidity Research. Microorganisms. 2020; 8(11):1643. https://doi.org/10.3390/microorganisms8111643
Chicago/Turabian StyleDenaro, Frank, Francesca Benedetti, Myla D. Worthington, Giovanni Scapagnini, Christopher C. Krauss, Sumiko Williams, Joseph Bryant, Harry Davis, Olga S. Latinovic, and Davide Zella. 2020. "The HIV-1 Transgenic Rat: Relevance for HIV Noninfectious Comorbidity Research" Microorganisms 8, no. 11: 1643. https://doi.org/10.3390/microorganisms8111643