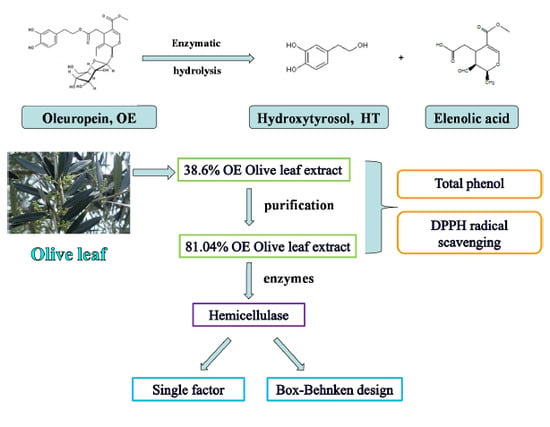
Enzymatic Hydrolysis of Oleuropein from Olea europea (Olive) Leaf Extract and Antioxidant Activities
Abstract
:1. Introduction

2. Results and Discussion
2.1. Preparation for High Purity OE

2.2. Enzymatic Hydrolysis of OE (Olive Leaf Extract) to HT
2.2.1. Enzyme Selection for HT Biotransformation

2.2.2. Substrate for HT Biotransformation

2.3. Enzyme Hydrolysis from High Purity OE (81.04%) Olive Leaf Extract to HT
2.3.1. Effect of pH

2.3.2. Effect of Temperature
2.3.3. Effect of Bioconversion Time
2.3.4. Effect of Enzyme Quantity
2.3.5. Optimization of Enzymatic Hydrolysis by RSM
| No. | X1 pH | X2 Temperature/°C | X3 Enzyme Quantity/mg | Y HT Content/% |
|---|---|---|---|---|
| 1 | −1 | −1 | 0 | 10.02 |
| 2 | 1 | −1 | 0 | 9.51 |
| 3 | −1 | 1 | 0 | 9.82 |
| 4 | 1 | 1 | 0 | 9.98 |
| 5 | −1 | 0 | −1 | 9.46 |
| 6 | 1 | 0 | −1 | 9.82 |
| 7 | −1 | 0 | 1 | 9.53 |
| 8 | 1 | 0 | 1 | 9.98 |
| 9 | 0 | −1 | −1 | 10.14 |
| 10 | 0 | 1 | −1 | 9.87 |
| 11 | 0 | −1 | 1 | 9.07 |
| 12 | 0 | 1 | 1 | 10.92 |
| 13 | 0 | 0 | 0 | 11.24 |
| 14 | 0 | 0 | 0 | 11.16 |
| 15 | 0 | 0 | 0 | 11.21 |
| 16 | 0 | 0 | 0 | 11.03 |
| 17 | 0 | 0 | 0 | 11.17 |
| Sources of Variation | Sum of Squares | df | Mean Square | F value | P value |
|---|---|---|---|---|---|
| model | 8.02 | 9 | 0.89 | 14.98 | 0.0009 |
| X1 | 0.026 | 1 | 0.026 | 0.44 | 0.5263 |
| X2 | 0.43 | 1 | 0.43 | 7.19 | 0.0315 |
| X3 | 0.00513 | 1 | 0.005513 | 0.093 | 0.7697 |
| X1X2 | 0.11 | 1 | 0.11 | 1.89 | 0.2120 |
| X1X3 | 0.00202 | 1 | 0.00202 | 0.034 | 0.8589 |
| X2X3 | 1.12 | 1 | 1.12 | 18.89 | 0.0034 |
| X12 | 2.80 | 1 | 2.80 | 47.13 | 0.0002 |
| X22 | 1.11 | 1 | 1.11 | 18.66 | 0.0035 |
| X32 | 1.77 | 1 | 1.77 | 29.77 | 0.0009 |
| residual | 0.42 | 7 | 0.059 | ||
| lack of fit | 0.39 | 3 | 0.13 | 20.12 | 0.0071 |
| pure error | 0.026 | 4 | 0.00647 | ||
| total | 8.43 | 16 | |||
| R2 | 0.9506 | ||||
| Radj2 | 0.8872 | ||||
| Adeq precision | 10.141 |
| Sources of Variation | Sum of Squares | df | Mean Square | F Value | P Value |
|---|---|---|---|---|---|
| model | 8.40 | 11 | 0.76 | 113.34 | <0.0001 |
| X1 | 0.16 | 1 | 0.16 | 24.34 | 0.0043 |
| X2 | 0.62 | 1 | 0.62 | 92.62 | 0.0002 |
| X3 | 0.005513 | 1 | 0.005513 | 0.82 | 0.4072 |
| X1X2 | 0.11 | 1 | 0.11 | 16.65 | 0.0095 |
| X1X3 | 0.002025 | 1 | 0.002025 | 0.30 | 0.6071 |
| X2X3 | 1.12 | 1 | 1.12 | 166.74 | <0.0001 |
| X12 | 2.80 | 1 | 2.80 | 416.06 | <0.0001 |
| X22 | 1.11 | 1 | 1.11 | 164.76 | <0.0001 |
| X32 | 1.77 | 1 | 1.77 | 262.78 | <0.0001 |
| X12 X2 | 0.21 | 1 | 0.21 | 31.83 | 0.0024 |
| X1 X22 | 0.17 | 1 | 0.17 | 24.96 | 0.0041 |
| residual | 0.034 | 5 | 0.006739 | ||
| lack of fit | 0.007812 | 1 | 0.007812 | 1.21 | 0.3335 |
| pure error | 0.026 | 4 | 0.00647 | ||
| total | 8.43 | 16 | |||
| R2 | 0.9960 | ||||
| Radj2 | 0.9872 | ||||
| Adeq precision | 29.880 |

2.4. Antioxidant Activity
| Samples | Phenol/% | HT/% | IC50/μg·mL−1 |
|---|---|---|---|
| 38.6% OE sample | 1.46 ± 0.03 | 0.25 ± 0.01 | 82.31 ± 1.26 |
| 38.6% OE sample enzymatic hydrolysate | 0.38 ± 0.01 | 2.81 ± 0.08 | 382.83 ± 0.72 |
| 81.04% OE sample | 2.96 ± 0.07 | 3.88 ± 0.08 | 9.43 ± 0.08 |
| 81.04% OE sample enzymatic hydrolysate 1 | 0.46 ± 0.01 | 5.64 ± 0.03 | 216.40 ± 0.54 |
| 81.04% sample ethyl acetate extracted enzymatic hydrolysate 2 | 2.89 ± 0.06 | 19.36 ± 0.31 | 41.82 ± 0.17 |
| BHT | 30.27 ± 0.55 | ||
| Vc | 3.31 ± 0.06 | ||
| OE | 4.97 ± 0.08 | ||
| HT | 1.07 ± 0.02 |
3. Experimental Section
3.1. Materials
| Enzymes | pH | Temperature/°C | Enzyme Activities/u·mg−1 | Form |
|---|---|---|---|---|
| β-glucosidase | 5 | 37 | 30 | powder |
| hemicellulase | 4.0–5.5 (5) | 45–60 (60) | 20 | powder |
| tannase | 4.5–6 (5) | 45–50 (50) | 10 | powder |
| neutral protease | 5.5–8 (7) | 45–50 (50) | 13 | powder |
| cellulase | 6–7 (6) | 45 | 10 | powder |
| glucoamylase | 4.0–4.5 (4.5) | 60 | 5 | powder |
| papain | 6–7 (6) | 55–65 (60) | 80 | powder |
| alkaline protease | 8 | 50 | 20 | powder |
| amylase | 4.2 | 60 | 3 | powder |
| β- glucanase | 6.0–6.5 (6) | 50–55 (50) | 20 | powder |
3.2. Preparation of the High Purity OE
3.2.1. Pretreatment of Resins
3.2.2. Saturated Static Adsorption
3.2.3. Static Adsorption Kinetics
3.2.4. Static Desorption for Different Concentrations of the Eluent
3.2.5. Dynamic Adsorptions
3.3. Enzymatic Hydrolysis
3.3.1. Enzyme Selection for HT Biotransformation
3.3.2. Substrate for HT Biotransformation
3.3.3. Single Factor Experimental Design
3.3.4. Box-Behnken Design
| Independent Variables | Symbol | Variable Levels | ||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| pH | X1 | 4.5 | 5.0 | 5.5 |
| Temperature (°C) | X2 | 40 | 50 | 60 |
| Enzyme quantity (mg) | X3 | 25 | 50 | 75 |
3.4. High Performance Liquid Chromatography (HPLC) Analysis
3.5. Total Phenol Content Determination
3.6. DPPH (1,1-Diphenyl-2-Picrylhydrazyl) Radical Scavenging Assay
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Abbreviation
| HT | hydroxytyrosol |
| OE | oleuropein |
| BHT | butylated hydroxytoluene |
| DPPH | 1,1-diphenyl-2-picrylhydrazyl |
| BG | β-glucosidase |
| T | tyrosol |
| CHD | coronary heart disease |
Conflicts of Interest
References
- Somova, L.I.; Shode, F.O.; Ramnanan, P.; Nadar, A. Antihypertensive, antiatherosclerotic and antioxidant activity of triterpenoids isolated from Olea europaea, subspecies africana leaves. J. Ethnopharmacol. 2003, 84, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Skerget, M.; Kotnik, P.; Hadolin, M.; Hras, A.R.; Simonic, M.; Knez, Z. Phenols, proanthocyanidins, flavones and flavonols in some plant materials and their antioxidant activities. Food Chem. 2005, 89, 191–198. [Google Scholar] [CrossRef]
- Sudjana, A.N.; D’Orazio, C.; Ryan, V.; Rasool, N.; Ng, J.; Islam, N.; Riley, T.V.; Hammer, K.A. Antimicrobial activity of commercial Olea europaea (olive) leaf extract. Int. J. Antimicrob. Agents 2009, 33, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.H.; Lee, B.Y. Antioxidant and antimicrobial activities of individual and combined phenolics in Olea europaea leaf extract. Bioresour. Technol. 2010, 101, 3751–3754. [Google Scholar] [CrossRef] [PubMed]
- Khayyal, M.T.; El-Ghazaly, M.A.; Abdallah, D.M.; Nassar, N.N.; Okpanyi, S.N.; Kreuter, M.H. Blood pressure lowering effect of an olive leaf extract (Oleaeuropaea) in L-NAME induced hypertension in rats. Arzneim.-Forsch. 2002, 52, 797–802. [Google Scholar]
- Fabiani, R.; de Bartolomeo, A.; Rosignoli, P. Virgin olive oil phenols inhibit proliferation of human promyelocytic leukemia cells (HL60) by inducing apoptosis and differentiation. J. Nutr. 2006, 136, 614–619. [Google Scholar] [PubMed]
- Bernini, R.; Crisante, F.; Merendino, N. Synthesis of a novel ester of hydroxytyrosol and α-lipoic acid exhibiting an antiproliferative effect on human colon cancer HT-29 cells. Eur. J. Med. Chem. 2011, 46, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Bouallagui, Z.; Han, J.; Isoda, H.; Sayadi, S. Hydroxytyrosol rich extract from olive leaves modulates cell cycle progression in MCF-7 human breast cancer cells. Food Chem. Toxicol. 2011, 179–184. [Google Scholar] [CrossRef]
- Lucia, D.M.; Panzella, L.; Pezzella, A.; Napolitano, A.; D’lschia, M. Oxidative chemistry of the natural antioxidant hydroxytyrosol: Hydrogen peroxide-dependent hydroxylation and hydroxyquinone/o-quinone coupling pathways. Tetrahedron 2006, 62, 1273–1278. [Google Scholar] [CrossRef]
- Bovicelli, P.; Antonioletti, R.; Mancini, S. Expedient synthesis of hydroxytyrosol and its esters. Synth. Commun. 2007, 37, 4245–4252. [Google Scholar] [CrossRef]
- Baraidi, P.G.; Simoni, D.; Manfredini, S. Preparation of 3,4-dihydroxy-1-benzeneethanol: A reinvestigation. Liebigs. Annal. Chem. 1983, 1983, 684–686. [Google Scholar] [CrossRef]
- Bai, C.; Yan, X.J.; Takenaka, M. Determination synthetic hydroxytyrosol in rat plasma by GC-MS. Agric. Food Chem. 1998, 46, 3998–4001. [Google Scholar] [CrossRef]
- Tuck, K.L.; Tan, H.W.; Hayball, P.J. Synthesis of tritiumlabeled hydroxytyrosol, a phenolic compound found in olive oil. Agric. Food Chem. 2000, 48, 4087–4090. [Google Scholar] [CrossRef]
- Yan, Y.; Guo, Y.; Peng, M.J.; An, J.; En, D. Synthesis of hydroxytyrosol. Fine Chem. 2012, 29, 985–988. [Google Scholar]
- Xu, C.; Gao, J.C.; Zhang, J.; Jiang, S.D. Process research of hydroxytyrosol synthesis. Fine Chem. 2010, 27, 1209–1212. [Google Scholar]
- Zhang, Z.L.; Chen, J.L.; Xu, Q.M. Effcient synthesis of hydroxytyrosol from 3,4-dihydroxybenzaldehyde. Synth. Commun. 2012, 42, 794–798. [Google Scholar] [CrossRef]
- Ye, J.Z.; Wang, C.Z.; Chen, H.X.; Zhou, H. Variation rule of hydroxytyrosol content in olive leaves. Chem. Ind. For. Prod. 2011, 31, 69–74. [Google Scholar]
- Wang, C.Z.; Gao, C.X.; Ye, J.Z.; Chen, X.J.; Jiang, W.Y. Study on seasonal variation of oleuropein content in olive leaves by HPLC. Chem. Ind. For. Prod. 2008, 28, 39–43. [Google Scholar]
- Briante, R.; Cara, F.L.; Tonziello, M.P.; Febbraio, F.; Nucci, R. Antioxidant Activity of the Main Bioactive Derivatives from Oleuropein Hydrolysis by Hyperthermophilic β-Glycosidase. J. Agric. Food Chem. 2001, 49, 3198–3203. [Google Scholar] [CrossRef] [PubMed]
- Briante, R.; Patumi, M.; Febbraio, F.; Nucci, R. Production of highly purified hydroxytyrosol from Olea europaea leaf extract biotransformed by hyperthermophilic β-glycosidase. J. Biotechnol. 2004, 111, 67–77. [Google Scholar]
- Espin, J.C.; Solar-Rivas, C.; Cantos, E. Synthesis of the antioxidant hydroxytyrosol using tyrosinase as biocatalyst. J. Agric. Food Chem. 2001, 49, 1187–1193. [Google Scholar] [CrossRef] [PubMed]
- Allouche, N.; Damak, M.; Ellouz1, R.; Sayadi, S. Use of Whole Cells of Pseudomonas aeruginosa for Synthesis of the Antioxidant Hydroxytyrosol via Conversion of Tyrosol. Appl. Environ. Microbiol. 2004, 70, 2105–2109. [Google Scholar] [CrossRef] [PubMed]
- Allouche, N.; Sayadi, S. Synthesis of hydroxytyrosol, 2-hydroxyphenylacetic acid, and 3-hydroxyphenylacetic acid by differential conversion of tyrosol isomers using Serratia Marcescens Strain. J. Agric. Food Chem. 2005, 53, 6525–6530. [Google Scholar] [CrossRef] [PubMed]
- Khoufi, S.; Hamza, M.; Sayadi, S. Enzymatic hydrolysis of olive wastewater for hydroxytyrosol enrichment. Bioresour. Technol. 2011, 102, 9050–9058. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, O.; Navarro, D.; Roche, M.; Asther, M.; Haon, M.; Delattre, M.; Lorquin, J.; Labat, M.; Asther, M.; Lesage-Meessen, L. Fungal enzymes as a powerful tool to release simple phenolic compounds from olive oil by-product. Process. Biochem. 2005, 40, 1855–1862. [Google Scholar] [CrossRef]
- Xue, Z.; Guo, Y.R.; Fu, C.C.; Huo, T.B.; Yan, M.; Sun, D.D. Extration of dietary fiber from apple flesh pomace using hemicellulase. Nat. Prod. Res. Dev. 2013, 25, 1474–1479. [Google Scholar]
- Rigane, G.; Bouaziz, M.; Baccar, N. Recovery of hydroxytyrosol rich extract from two-phase chemlali olive pomace by chemical treatment. J. Food Sci. 2012, 77, 1077–1083. [Google Scholar] [CrossRef]
- Bu, W.W.; Liu, C.J.; Tian, S.F. Comparison of extraction of olive leaves to prepare hydroxytyrosol by hydrochloric acid and β-glycosidase. Sci. Technol. Food Ind. 2011, 32, 228–232. [Google Scholar]
- Liu, Q.; Li, Y.; Yao, H.Y. Determination of total polyphenol in barley seed extracts by Folin-Ciocalteu colorimetry. Food Sci. Technol. 2007, 4, 175–177. [Google Scholar]
- Ni, Q.X.; Xu, G.Z.; Wang, Z.Q. Seasonal variations of the antioxidant composition in ground bamboo Sasa argenteastriatus leaves. Int. J. Mol. Sci. 2012, 13, 2249–2262. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, J.-J.; Wang, C.-Z.; Ye, J.-Z.; Tao, R.; Zhang, Y.-S. Enzymatic Hydrolysis of Oleuropein from Olea europea (Olive) Leaf Extract and Antioxidant Activities. Molecules 2015, 20, 2903-2921. https://doi.org/10.3390/molecules20022903
Yuan J-J, Wang C-Z, Ye J-Z, Tao R, Zhang Y-S. Enzymatic Hydrolysis of Oleuropein from Olea europea (Olive) Leaf Extract and Antioxidant Activities. Molecules. 2015; 20(2):2903-2921. https://doi.org/10.3390/molecules20022903
Chicago/Turabian StyleYuan, Jiao-Jiao, Cheng-Zhang Wang, Jian-Zhong Ye, Ran Tao, and Yu-Si Zhang. 2015. "Enzymatic Hydrolysis of Oleuropein from Olea europea (Olive) Leaf Extract and Antioxidant Activities" Molecules 20, no. 2: 2903-2921. https://doi.org/10.3390/molecules20022903